













| General Information: |
|
| Name(s) found: |
TOM1 /
YDR457W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 3268 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA nucleolus [IDA |
| Biological Process: |
nucleocytoplasmic transport
[IMP protein polyubiquitination [TAS regulation of cell size [IMP mitosis [IMP protein monoubiquitination [TAS nucleus organization [IMP |
| Molecular Function: |
ubiquitin-protein ligase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLFTRCEKA RKEKLAAGYK PLVDYLIDCD TPTFLERIEA IQEWDRSRDD LYVWIPILDR 60
61 MDGLLLKVAE KYKYKQDPKK ECEVKLVEME AHDVDYCLKM LKFTRRLLLN TENRFVYSSG 120
121 DVLMYLLNCP NFTIKLAVMR ILAILGERFV IAREKIVAHN IFGDHNLRKK TLKLALSLSS 180
181 SVMDEDGEHF SLVDLYFDKK KVPQKWRKLR FTHYTSNDFK KSSQQKNNIN ETQTSIKKVT 240
241 MTTQELCEHS LQQIFDKGMA LLPAESWFDF SIKASVAKAF SDDSGENIDL RNIIIETKLN 300
301 AIAFVNTIFS PPQVSSKLFE LDPYAFNSLT DLISLSETKI PKELRTDALF TLECISLKHV 360
361 WCSDIIRNLG GNISHGLLFQ ILRYIAKTLR EATDEIDEEY NVRFFYLISN LADVKPLHES 420
421 LFAAGLIPTL LEIVSIRNCP YKRTLASATH LLETFIDNSE TTTEFIENDG FTMLITSVAN 480
481 EIDFTLAHPE TWQPPKYSVV YYSISFRELA YIRSLLKLVL KLLSTDSGDR IRNLIDSPIL 540
541 VSLKKILENK LVFGLTLITY TLDVVQKVIN SEPTIYPVLV EAGLIPYVID NFPKLIGPSA 600
601 ELLSLLPDVV SAICLNPEGL KQVKEKGLIN NLFDFLLDAD HARILTGGDR STEYGTDIDE 660
661 LARHYPDLKA NIVEALCNVI RKMPSTFRNE REFLFTSPKD QKYFFHRKNE EILTDKEEHE 720
721 PAYWELLDKG TMLDTFTSVL FGMSLGNGSF SQVPQHLEAR DFLAIIFMEN PPYEYFTSVA 780
781 ISNVTEVLQY LDEKYEDYAF MDVMKVLNDQ LENLNDFLNS PNDRSFFLER DGENSVRSCH 840
841 SKLCRLAAIL NIVTNVYIDL TTLSCKRIMQ IYSYFDKRGF SLIKNLKLLF QKCALEEMYI 900
901 RQHMPDSVIT ETMPLPIVDV SGDGPPLQIY IDDPKKGDQK GKITSVKTRN TLQMRTILYT 960
961 LQSNTAILFR CFLRLSHSRN MDLEHKDLTT EVHIFENVVE NVIEMLKATE LEGHLPYILV1020
1021 LLNFNTFVFT IPKASPNSTE ILQTIPAYIF YQKGGYLLYL HIIRDLFTRM TKIKDLSSLD1080
1081 NINYIDESNG ILTLSCLINA LTFYNKSMQT ETMENVQSIG KYYVSIDDDY NIMKALTVPI1140
1141 KVMALAMILD LDKSDSLFKT QSRNVPYSVF KQLLSMLKNI FTNVNIYTKE LYELHWDLIF1200
1201 PPIKKISLFE QVGIPGDVAA NYLTDTGDDL PADNSIGLFS PEQWEKYKKL IGEDKSIYYP1260
1261 QPMQAQYYKG CSSKELDELR DTFFNDGLPS RIFTVLPFYP KLVNAFAKTL LQIFTKYDEP1320
1321 TEVFAGRILD RILETDLDDP ATLSSLIHLF GIFLNEKYIY QKASHLMQRF IEYLEKSLKP1380
1381 EHVNTPWFSK ALYVYEIILA KSELPHLEEL SKDVLLRYPL LSMAKVFRIP DPMKQKLFDI1440
1441 LIRVSDISNF YSALATSRIL IFYSRDELYA NNIARSGILS RLLKVIGSFQ KLDKINFLES1500
1501 SFLLLTRRCF ETTENVDALI RAEINRSFTA RPLGGGDDAV RELTTILEEK AHVVMRSPSQ1560
1561 FIDVLCETAR FHEFDDQGAL VDYSLKRFLG EKDKNTQASS TEKSDIYERT GIMHLLLSQL1620
1621 MAASEKDWLS EPANSSDLPE NKKAQLDPSR NPVCAYMIFL LKLLVELVSS YNQCKFEFLT1680
1681 FSRRNTYAER PRPRTTAINF FLYRLLDKPV GTDHDKHEAK RREVIGMLAR SVIIGFLATV1740
1741 QDDRTTKTDV KLADPHMNFI RKFAIEAIIK AIRNATSSSK LLESNHLKLD MWFRIITSMV1800
1801 YVQAPYLRQL LDSNKVEADQ YQLCKLVIDL GLPSVITEAM ASIDLNYPFS KKIFNVAVEA1860
1861 LNTISSTRNN FSEHFKIEDH DEVEDEVDES DKEEIPDMFK NSALGMYDVE DIEEDDDDDT1920
1921 SLIGDDDAMA FVDSDNGFEV VFSDEDDDMG EEDADDARSD SEENELSSEM QSSTADGTDV1980
1981 DYEVDDADGL IINIDQPSGD DEEMADYDAN ISHSSHSENE DDASMDVIEV YDDELSSGYD2040
2041 VDLSDYDVDE SDWDSGLSSL SISDEDSESS EDEPINSTRM GDSRRRWLIA EGVELTDDSQ2100
2101 GESEEDDRGV FRGIEHIFSN ENEPLFRVHD EMRHRNHHRS INRTHFHSAM SAPSLSLLNR2160
2161 GRRNQSNLIN PLGPTGLEQV ENDISDQVTV AGSGSRPRSH HLHFSEVLVS GSFFDEPVLD2220
2221 GIILKSTVSR WKDIFDMFYD SKTYANCIIP TVINRLYKVS LALQKDLENK REQEKLKNKN2280
2281 LLFNEAKVES HNSSDAISVE QDDIQESNVT HDDHEPVYVT IQGSEVDIGG TDIDPEFMNA2340
2341 LPDDIRADVF AQHVRERRAE ARLNSDHNVH SREIDSDFLE AIPEDIREGI LDTEAEEQRM2400
2401 FGRIGSSADV IRADDDVSNN DEEVENGLDH GNSNDRNNAD PEKKKPARIY FAPLIDRAGI2460
2461 ASLMKSVFIS KPYIQREIYH ELFYRLCSSK QNRNDLMNTF LFILSEGIID QHSLEKVYNI2520
2521 ISSRAMGHAK TTTVRQLPSD CTPLTVANQT IEILQSLIDA DSRLKYFLIA EHDNLIVNKA2580
2581 NNKSRKEALP DKKLRWPLWH LFSLLDRKLI TDESVLMDLL TRILQVCTKT LAVLSTSSNG2640
2641 KENLSKKFHL PSFDEDDLMK ILSIIMLDSC TTRVFQQTLN IIYNLSKLQG CMSIFTKHLV2700
2701 SLAISIMSKL KSALDGLSRE VGTITTGMEI NSELLQKFTL PSSDQAKLLK ILTTVDFLYT2760
2761 HKRKEEERNV KDLQSLYDKM NGGPVWSSLS ECLSQFEKSQ AINTSATILL PLIESLMVVC2820
2821 RRSDLSQNRN TAVKYEDAKL LDFSKTRVEN LFFPFTDAHK KLLNQMIRSN PKLMSGPFAL2880
2881 LVKNPKVLDF DNKRYFFNAK LKSDNQERPK LPITVRREQV FLDSYRALFF KTNDEIKNSK2940
2941 LEITFKGESG VDAGGVTREW YQVLSRQMFN PDYALFLPVP SDKTTFHPNR TSGINPEHLS3000
3001 FFKFIGMIIG KAIRDQCFLD CHFSREVYKN ILGRPVSLKD MESLDPDYYK SLVWILENDI3060
3061 TDIIEETFSV ETDDYGEHKV INLIEGGKDI IVTEANKQDY VKKVVEYKLQ TSVKEQMDNF3120
3121 LVGFYALISK DLITIFDEQE LELLISGLPD IDVDDWKNNT TYVNYTATCK EVSYFWRAVR3180
3181 SFDAEERAKL LQFVTGTSKV PLNGFKELSG VNGVCKFSIH RDFGSSERLP SSHTCFNQLN3240
3241 LPPYESYETL RGSLLLAINE GHEGFGLA |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | 3912: TAP-tagged, strain background includes hpc2(delta) | Green EM, et al (2005) | |
| View Run | 3829 - low filter | Green EM, et al (2005) | |
| View Run | Sample 2- cpn2 from june 2005 | McCusker D, et al (2007) | |
| View Run | Sample boi 1 with ha tag from october 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
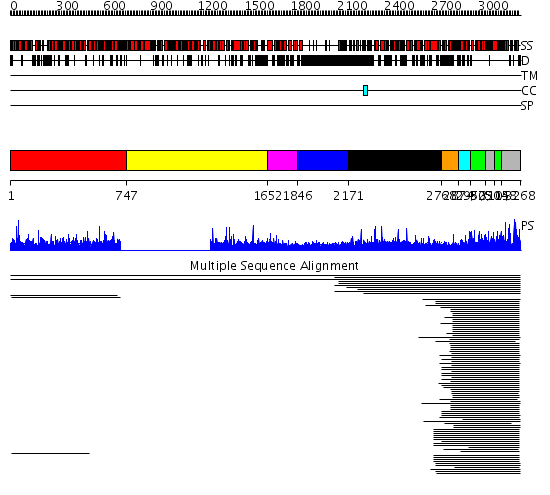
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..746] | 2.005001 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [747..1651] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [1652..1845] | 1.30299 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1846..2170] | 29.460997 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [2171..2767] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [2768..2873] | 2.30103 | Ubiquitin ligase NEDD4 WWIII domain | |
| 7 | View Details | [2874..2951] | 578.9691 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 8 | View Details | [2952..3049] [3105..3147] |
578.9691 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 9 | View Details | [3050..3104] [3148..3268] |
578.9691 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)