













| General Information: |
|
| Name(s) found: |
RFA2 /
YNL312W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 273 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chromosome, telomeric region
[IMP DNA replication factor A complex [TAS |
| Biological Process: |
DNA strand elongation involved in DNA replication
[TAS double-strand break repair via homologous recombination [IGI DNA recombination [TAS nucleotide-excision repair [TAS DNA unwinding involved in replication [TAS postreplication repair [TAS DNA replication, synthesis of RNA primer [TAS |
| Molecular Function: |
DNA binding
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATYQPYNEY SSVTGGGFEN SESRPGSGES ETNTRVNTLT PVTIKQILES KQDIQDGPFV 60
61 SHNQELHHVC FVGVVRNITD HTANIFLTIE DGTGQIEVRK WSEDANDLAA GNDDSSGKGY 120
121 GSQVAQQFEI GGYVKVFGAL KEFGGKKNIQ YAVIKPIDSF NEVLTHHLEV IKCHSIASGM 180
181 MKQPLESASN NNGQSLFVKD DNDTSSGSSP LQRILEFCKK QCEGKDANSF AVPIPLISQS 240
241 LNLDETTVRN CCTTLTDQGF IYPTFDDNNF FAL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
DNA2 MPH1 RAD52 RFA1 RFA2 RFA3 RIM1 RTT105 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
RFA1 RFA2 RFA3 |
| View Details | Krogan NJ, et al. (2006) |

|
DNA2 HSP12 PRO1 RFA1 RFA2 RFA3 |
| View Details | Gavin AC, et al. (2002) |

|
CMR1 DNA2 ECM29 HDA1 HDA2 HDA3 HSC82 KAP95 LCD1 MEC1 MGM101 MLH1 MPH1 MRPS28 MSH2 MSH3 MSH6 OLA1 PGK1 PMS1 PRO2 RAD16 RAD23 RAD34 RAD4 RAD52 RFA1 RFA2 RFA3 RIM1 RNH1 RPN10 RPN12 RPN3 RPN6 RPN8 RPN9 RPT2 RPT3 RPT6 RSC8 RVB2 SGS1 SHS1 SRO7 SSL1 TFA1 TFB1 TFB4 TIF1, TIF2 TOP3 VAS1 VPS1 YHR122W |
| View Details | Ho Y, et al. (2002) |

|
AHP1 CDC10 GCD11 HIR3 HTB1 MGM101 RFA2 |
| View Details | Qiu et al. (2008) |

|
MCM4 MCM6 PRI2 RFA1 RFA2 RFA3 RFC3 RFC4 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
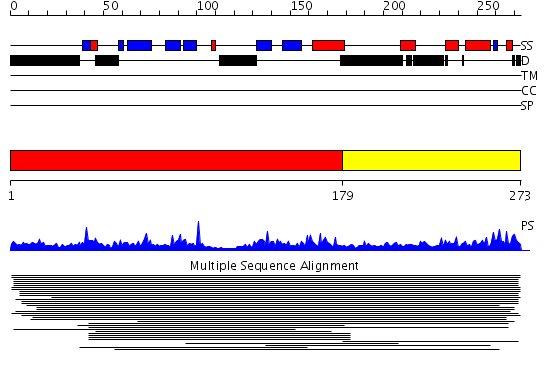
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..178] | 142.69897 | Replication protein A 32 KDa subunit (RPA32) fragment | |
| 2 | View Details | [179..273] | 25.69897 | C-terminal domain of RPA32 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)