













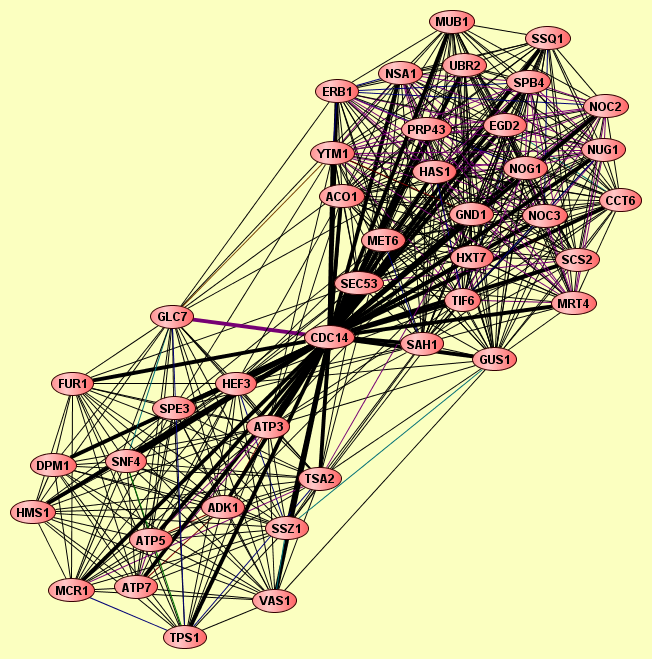
| Protein: | CDC14 |
| From Publication: | Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002 Jan 10;415(6868):180-3. |
| Complexes containing CDC14: | 2 |
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 17 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Cdc14. The topolgies of protein complexes in this experiment are unknown. | ADK1 ATP3 ATP5 ATP7 CDC14 DPM1 FUR1 GLC7 HEF3 HMS1 MCR1 SNF4 SPE3 SSZ1 TPS1 TSA2 VAS1 |
| View GO Analysis | 26 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Erb1. The topolgies of protein complexes in this experiment are unknown. | ACO1 CCT6 CDC14 EGD2 ERB1 GND1 GUS1 HAS1 HXT7 MET6 MRT4 MUB1 NOC2 NOC3 NOG1 NSA1 NUG1 PRP43 SAH1 SCS2 SEC53 SPB4 SSQ1 TIF6 UBR2 YTM1 |