













| General Information: |
|
| Name(s) found: |
SKI3 /
YPR189W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1432 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS cytoplasm [IDA |
| Biological Process: |
response to exogenous dsRNA
[IMP mRNA catabolic process [IMP |
| Molecular Function: |
translation repressor activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDIKQLLKE AKQELTNRDY EETIEISEKV LKLDPDNYFA HIFLGKALSS LPASNNVSSN 60
61 RNLERATNHY VSAAKLVPDN LLAWKGLFLL FRTTEVVPDI LSYDEYFDLC GQYADALLKQ 120
121 EQSQVELIND IKLLKKTHPD CQKAFYQHLK PGSLMAETIG RHLSTPQDAL LNLIKILSNI 180
181 ETTEIGKTLS QNRLKLKASD PDYQIKLNSF SWEIIKNSEI DQLYNQLVNI LADDQKRSEI 240
241 ENQWLEYRIK VLKSMPLDVK KDFFTKVKEM VEDMVLVNHQ SLLAWQKYFE WTDYEDLDNM 300
301 DAPLIIKYFK KFPKDPLAMI LYSWLSSKLS KYDIKSLESA NKPPEGHKKT EKETDIKDVD 360
361 ETNEDEVKDR VEDEVKDRVE DEVKDQDEEA KEDEEEDLDD IEIGLLEEEV VTVLTENIVK 420
421 CKNNILAHRI LCQYYLLTKE YEAALPYIKN GISLIAYNIK DLGVHLPLTK REFSLDLATV 480
481 YTYVDAPKDH NAALKLYDNI LSGDFSNIQA KMGKGIIFIE RKNWKDAMTL LTQVHEQSPN 540
541 NLEVLSELSW SKAHMGYMDE ALAGLDTVIK GIKGMDLRSI DFRALNLWRQ AKVYIMKHAS 600
601 INDAKQENVK CAFKLLIQSI KILDTFAPGF STLGDIYCHY YKDHLRAFKC YFKAFDLDAG 660
661 DYTAAKYITE TYASKPNWQA ASSIASRLIK GEKAKAELRS NNWPFRVVGI AHLEKQEESD 720
721 SIEWFQSALR VDPNDVESWV GLGQAYHACG RIEASIKVFD KAIQLRPSHT FAQYFKAISL 780
781 CDVGEYLESL DILEKVCQEA ATEESFQIGL VEVLMRCSLD LYSQGFLLKS VSIAKDTIER 840
841 IKIIISELKC ENQQVWIYLS QVLRLFIWIE SKVDTLPVES LVSIFENSQF SGSEEIDSVD 900
901 NIKIDTLLDS TTDDNVSIAC KFLILASKYS VSDQKFTDIA GTVRASYWYN IGISELTAFI 960
961 TLKEPQYRDA AIFAFKKSIQ LQSNTSETWI GLGIATMDIN FRVSQHCFIK ATALEPKATN1020
1021 TWFNLAMLGL KKKDTEFAQQ VLNKLQSLAP QDSSPWLGMA LILEEQGDII GSSKLFAHSF1080
1081 ILSNGRSKAA QFMYAKNVLE NHINNGDDER DIETVEKLTT ASIALEQFFK KSPDSQFALQ1140
1141 CALLTLERLH HYENANELAN RLIGILEKKF EKTQDERELF NFAIIKGQFA RIHLGLGNFE1200
1201 LSIENADLSQ GIISESSDEK SMKTKISNHI CLGLSYFFLN DFDQTLNQFQ ELLSISKDSK1260
1261 HLVVLIAKVL YDVGESDTKE IALQELTEYI ATSGADLLVT LTIAAMSILD DKREDLSIIL1320
1321 EELKALPLSK QIIDKHKDAP YLIEEITKRL YRNDTGKQVW QRSAYFFPNN LKVWERLDKN1380
1381 IQRRIASNGQ NKVTAEEMSK LYCESKNLRS IQRGMFLCPW NVTAVKALNE CF |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
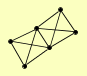
|
RRP43 SKI2 SKI3 SKI7 SKI8 YKL023W |
| View Details | Krogan NJ, et al. (2006) |
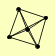
|
ENT5 SKI2 SKI3 SKI8 YKL023W |
| View Details | Gavin AC, et al. (2006) |
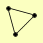
|
SKI2 SKI3 SKI8 |
| View Details | Gavin AC, et al. (2002) |
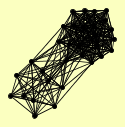
|
AMN1 ASC1 BEM2 BUD14 CSL4 DIS3 ECM16 KIN3 LRP1 MIS1 MTR3 RRP4 RRP40 RRP42 RRP43 RRP45 RRP46 RRP6 SCP160 SKI2 SKI3 SKI6 SKI7 SRP1 SSZ1 TIF4631 UTP10 UTP20 UTP22 YIR035C ZUO1 |
| View Details | Ho Y, et al. (2002) |
|
AKL1 SKI2 SKI3 SKI8 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
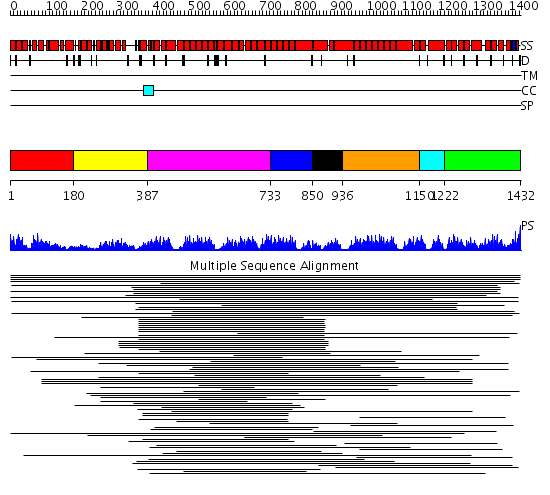
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..179] | 10.193996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [180..386] | 1.565997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [387..732] | 20.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 4 | View Details | [733..849] | 18.0 | Designed protein cTPR3 | |
| 5 | View Details | [850..935] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [936..1149] | 4.69897 | Protein farnesyltransferase alpha-subunit | |
| 7 | View Details | [1150..1221] | 7.39794 | Hop | |
| 8 | View Details | [1222..1432] | 9.537997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)