













| General Information: |
|
| Name(s) found: |
APC2 /
YLR127C
[SGD]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 853 amino acids |
Gene Ontology: |
|
| Cellular Component: |
anaphase-promoting complex
[TAS |
| Biological Process: |
mitotic spindle elongation
[TAS cyclin catabolic process [TAS mitotic sister chromatid segregation [TAS anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process [IPI mitotic metaphase/anaphase transition [TAS protein ubiquitination [IPI |
| Molecular Function: |
ubiquitin-protein ligase activity
[TAS protein binding [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFQITPTRD LKVITDELQT LSSYIFHTNI VDDLNSLLTW MSPNDAKSNH QLRPPSLRIK 60
61 NIIKVLFPNN ATTSPYSMIN TSQANNSIVN EGNTNKELQL QLFSTLKEFY IFQVRYHFFL 120
121 HFNNINYLKD IQRWENYYEF PLRYVPIFDV NVNDWALELN SLRHYLLNRN IKFKNNLRTR 180
181 LDKLIMDDDF DLADNLIQWL KSANGSLSST ELIVNALYSK INKFCEDNMS RVWNKRFMIM 240
241 ETFNKFINQY WSQFSKLVGC PEDDHELTTT VFNCFESNFL RIRTNEIFDI CVLAYPDSKV 300
301 TLLELRKIMK DFKDYTNIVT TFLSDFKKYI LNPSVTTVDA LLRYVKTIKA FLVLDPTGRC 360
361 LHSITTFVKP YFQERKHLVN VLLYAMLDLP EEELKEKINF NVDMKALLSL VDTLHDSDIN 420
421 QDTNITKRDK NKKSPFLWNL KVKGKRELNK DLPIRHAMLY EHILNYYIAW VPEPNDMIPG 480
481 NIKSSYIKTN LFEVLLDLFE SREFFISEFR NLLTDRLFTL KFYTLDEKWT RCLKLIREKI 540
541 VKFTETSHSN YITNGILGLL ETTAPAADAD QSNLNSIDVM LWDIKCSEEL CRKMHEVAGL 600
601 DPIIFPKFIS LLYWKYNCDT QGSNDLAFHL PIDLERELQK YSDIYSQLKP GRKLQLCKDK 660
661 GKVEIQLAFK DGRKLVLDVS LEQCSVINQF DSPNDEPICL SLEQLSESLN IAPPRLTHLL 720
721 DFWIQKGVLL KENGTYSVIE HSEMDFDQAQ KTAPMEIENS NYELHNDSEI ERKYELTLQR 780
781 SLPFIEGMLT NLGAMKLHKI HSFLKITVPK DWGYNRITLQ QLEGYLNTLA DEGRLKYIAN 840
841 GSYEIVKNGH KNS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
APC1 APC2 APC4 APC5 CDC16 CDC23 CDC27 DOC1 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
APC1 APC11 APC2 APC4 APC5 APC9 CDC16 CDC23 CDC26 CDC27 DOC1 |
| View Details | Krogan NJ, et al. (2006) |
|
APC1 APC2 APC4 APC5 CDC16 CDC23 CDC26 CDC27 DOC1 FMP52 MND2 SSP2 |
| View Details | Gavin AC, et al. (2006) |
|
APC1 APC2 APC4 APC5 APC9 CDC16 CDC23 CDC27 |
| View Details | Gavin AC, et al. (2002) |
|
ACT1 APC1 APC2 CDC16 CDC23 CDC27 |
| View Details | Ho Y, et al. (2002) |
|
APC2 FMP48 HIS4 |
| View Details | Qiu et al. (2008) |
|
APC1 APC11 APC2 APC4 APC5 APC9 BUL2 CDC16 CDC23 CDC26 CDC27 DOC1 SWM1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
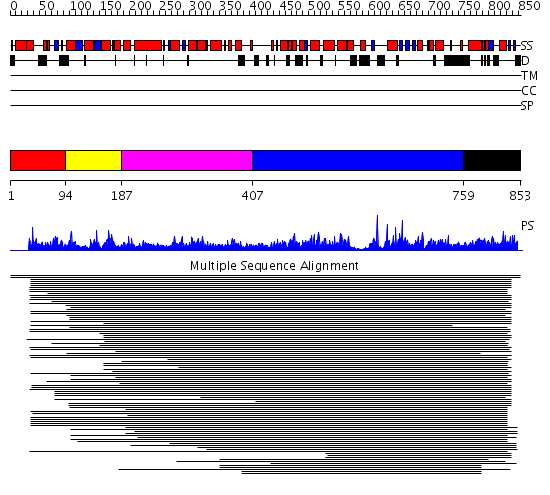
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..93] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [94..186] | 1.05099 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [187..406] | 2.071982 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [407..758] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [759..853] | 375.64593 | Anaphase promoting complex (APC) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)