













| General Information: |
|
| Name(s) found: |
DIG2 /
YDR480W
[SGD]
|
| Description(s) found:
Found 31 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 323 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[TAS |
| Biological Process: |
invasive growth in response to glucose limitation
[IGI |
| Molecular Function: |
transcription factor binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNKEEQEDPQ QEQISTVQEN DPRNLQQLGM LLVSPGLDED RLSEKMISKI KKSRDIEKNQ 60
61 KLLISRLSQK EEDHSGKPPT ITTSPAEKTV PFKSLNHSLK RKRVPPALNF SDIQASSHLH 120
121 GSKSAPPNIT RFPQHKNSLR VRYMGRMAPT NQDYHPSVAN SYMTATYPYP YTGLPPVPCY 180
181 PYSSTPTQTH AYEGYYSPMY PGPLYNNGII PADYHAKRKK LAGRSPHLED LTSRKRTFVS 240
241 KHHNGDPIIS KTDEDIECSV TKNSLSEGAS LNDDADDDND KERIIIGEIS LYDDVFKFEV 300
301 RDDKNDYMKA CETIWTEWHN LKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
DIG1 DIG2 KSS1 STE12 |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Ho Y, et al. (2002) |

|
ACO1 ARP7 AVO1 BEM3 CCT2 CYS4 DIG1 DIG2 FET4 GFA1 GUS1 HAS1 HXT6 KSS1 MKT1 MSE1 NAP1 NPA3 PHO84 PIM1 PMA1 PYC1 RGC1 RPA135 RPN10 RPN6 RVB1 SEN1 SRP1 STE11 STE12 STE7 TEC1 TSC11 UBI4 YDR239C YHR033W |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
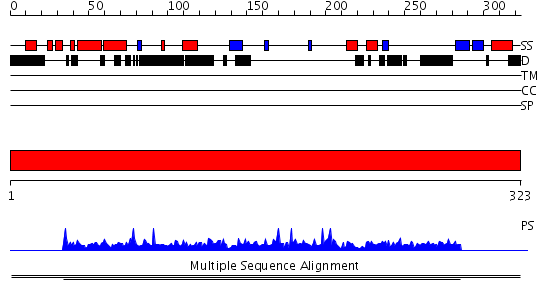
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..323] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [174..323] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)