













| General Information: |
|
| Name(s) found: |
MRPS9 /
YBR146W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 278 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA mitochondrial small ribosomal subunit [IDA |
| Biological Process: |
translation
[IDA |
| Molecular Function: |
structural constituent of ribosome
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFSRLSLFRR AALAPAPMRM SFRTIYQKTE DELPRRIVPK LATFYSANPN HEDRINRLER 60
61 LLRKYIKLPS QNNNEAQQTK APWISFDEYA LIGGGTKLKP TQYTQLLYML NKLHNIDPQL 120
121 TNDEITSELS QYYKKSSMLS NNIKIKTLDE FGRSIAVGKR KSSTAKVFVV RGTGEILVNG 180
181 RQLNDYFLKM KDRESIMYPL QVIESVGKYN IFATTSGGGP TGQAESIMHA IAKALVVFNP 240
241 LLKSRLHKAG VLTRDYRHVE RKKPGKKKAR KMPTWVKR |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample 3 - cpn + atp, from june 2005 | McCusker D, et al (2007) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
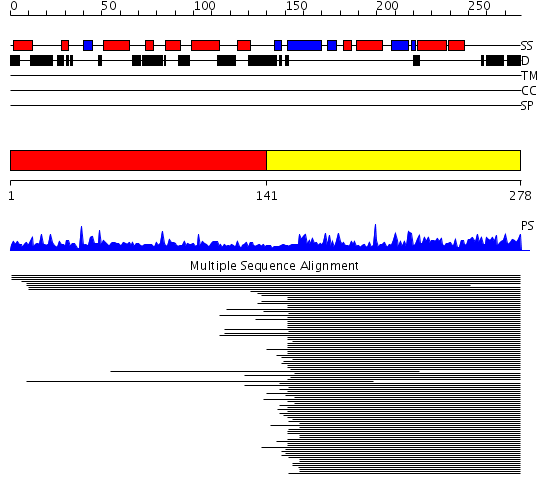
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..140] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [141..278] | 264.70927 | Ribosomal protein S9 |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)