













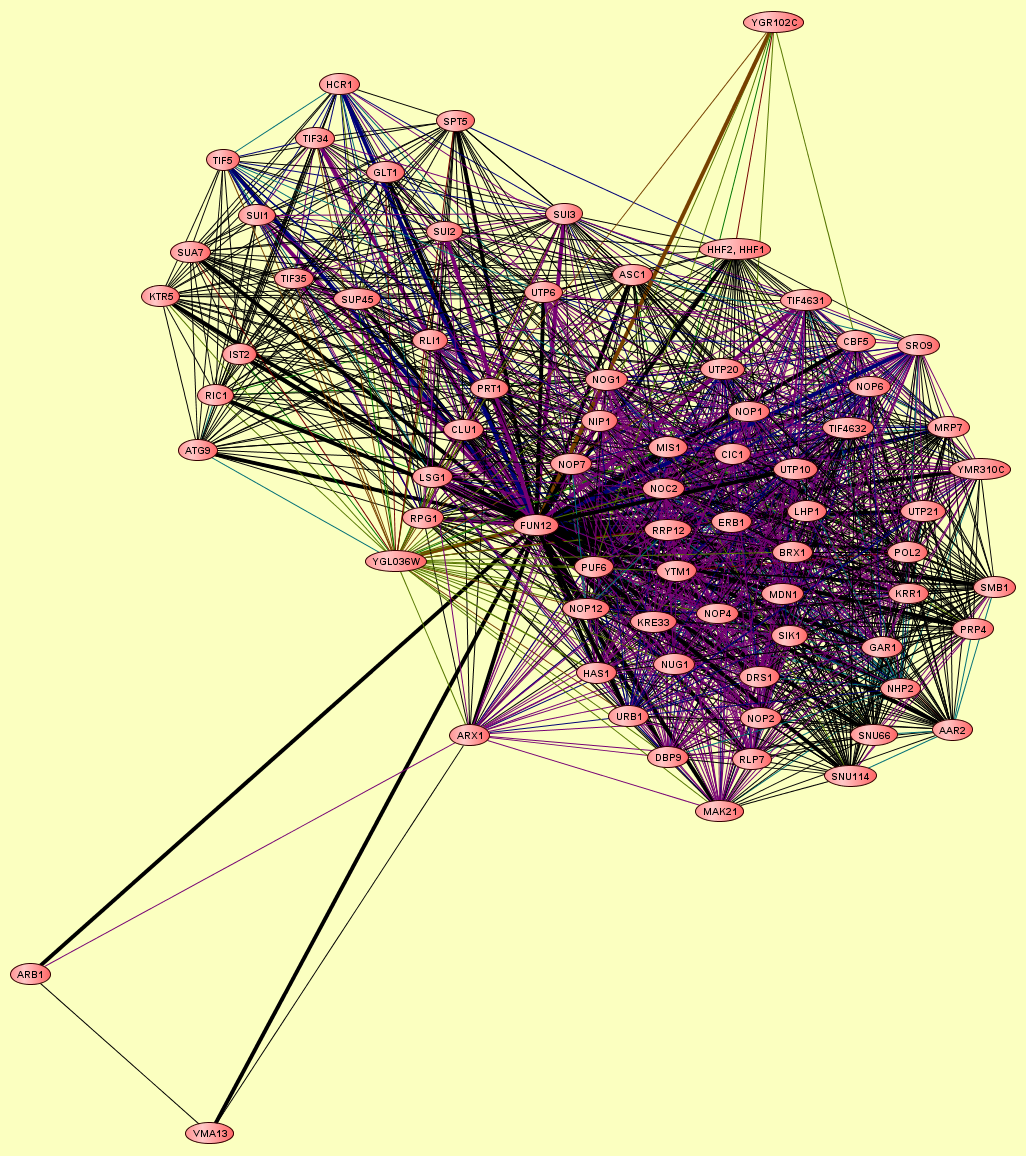
| Protein: | FUN12 |
| From Publication: | Gavin A.C. et al. (2002) Abstract Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002 Jan 10;415(6868):141-7. |
| Complexes containing FUN12: | 4 |
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 9 proteins | From the published set of protein complexes. | CBF5 FUN12 HHF1, HHF2 MIS1 NIP1 NOP1 PRT1 PUF6 YGR102C |
| View GO Analysis | 46 proteins | From the published set of protein complexes. | AAR2 BRX1 CBF5 CIC1 DBP9 DRS1 ERB1 FUN12 GAR1 HAS1 KRE33 KRR1 LHP1 MAK21 MDN1 MIS1 MRP7 NHP2 NIP1 NOC2 NOG1 NOP1 NOP12 NOP2 NOP4 NOP56 NOP6 NOP7 NUG1 POL2 PRP4 PUF6 RLP7 RRP12 SMB1 SNU114 SNU66 SRO9 TIF4631 TIF4632 URB1 UTP10 UTP20 UTP21 YMR310C YTM1 |
| View GO Analysis | 27 proteins | From the published set of protein complexes. | ASC1 ATG9 CLU1 FUN12 GLT1 HCR1 IST2 KTR5 LSG1 NIP1 NOG1 NOP7 PRT1 RIC1 RLI1 RPG1 SPT5 SUA7 SUI1 SUI2 SUI3 SUP45 TIF34 TIF35 TIF5 UTP6 YGL036W |
| View GO Analysis | 4 proteins | From the published set of protein complexes. | ARB1 ARX1 FUN12 VMA13 |