













| General Information: |
|
| Name(s) found: |
ISW2 /
YOR304W
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1120 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI chromatin accessibility complex [IPI |
| Biological Process: |
negative regulation of transcription from RNA polymerase II promoter by pheromones
[IMP chromatin remodeling [IGI nucleosome positioning [IDA chromatin silencing at telomere [IMP |
| Molecular Function: |
ATPase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTQQEEQRS DTKNSKSESP SEVLVDTLDS KSNGSSDDDN IGQSEELSDK EIYTVEDRPP 60
61 EYWAQRKKKF VLDVDPKYAK QKDKSDTYKR FKYLLGVTDL FRHFIGIKAK HDKNIQKLLK 120
121 QLDSDANKLS KSHSTVSSSS RHHRKTEKEE DAELMADEEE EIVDTYQEDI FVSESPSFVK 180
181 SGKLRDYQVQ GLNWLISLHE NKLSGILADE MGLGKTLQTI SFLGYLRYVK QIEGPFLIIV 240
241 PKSTLDNWRR EFLKWTPNVN VLVLHGDKDT RADIVRNIIL EARFDVLITS YEMVIREKNA 300
301 LKRLAWQYIV IDEAHRIKNE QSALSQIIRL FYSKNRLLIT GTPLQNNLHE LWALLNFLLP 360
361 DIFGDSELFD EWFEQNNSEQ DQEIVIQQLH SVLNPFLLRR VKADVEKSLL PKIETNVYVG 420
421 MTDMQIQWYK SLLEKDIDAV NGAVGKREGK TRLLNIVMQL RKCCNHPYLF EGAEPGPPYT 480
481 TDEHLIFNSG KMIILDKLLK RLKEKGSRVL IFSQMSRLLD ILEDYCYFRD FEYCRIDGST 540
541 SHEERIEAID EYNKPNSEKF VFLLTTRAGG LGINLVTADT VILFDSDWNP QADLQAMDRA 600
601 HRIGQKKQVH VYRFVTENAI EEKVIERAAQ KLRLDQLVIQ QGTGKKTASL GNSKDDLLDM 660
661 IQFGAKNMFE KKASKVTVDA DIDDILKKGE QKTQELNAKY QSLGLDDLQK FNGIENQSAY 720
721 EWNGKSFQKK SNDKVVEWIN PSRRERRREQ TTYSVDDYYK EIIGGGSKSA SKQTPQPKAP 780
781 RAPKVIHGQD FQFFPKELDA LQEKEQLYFK KKVNYKVTSY DITGDIRNEG SDAEEEEGEY 840
841 KNAANTEGHK GHEELKRRIE EEQEKINSAP DFTQEDELRK QELISKAFTN WNKRDFMAFI 900
901 NACAKYGRDD MENIKKSIDS KTPEEVEVYA KIFWERLKEI NGWEKYLHNV ELGEKKNEKL 960
961 KFQETLLRQK IEQCKHPLHE LIIQYPPNNA RRTYNTLEDK FLLLAVNKYG LRADKLYEKL1020
1021 KQEIMMSDLF TFDWFIKTRT VHELSKRVHT LLTLIVREYE QPDANKKKRS RTSATREDTP1080
1081 LSQNESTRAS TVPNLPTTMV TNQKDTNDHV DKRTKIDQEA |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
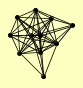
|
DPB4 HTA2 ISW1 ISW2 ITC1 MOT1 RFA1 RPO31 RSC8 STH1 VPS1 |
| View Details | Krogan NJ, et al. (2006) |
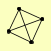
|
DLS1 ELA1 ISW2 ITC1 |
| View Details | Gavin AC, et al. (2002) |

|
ACT1 ADE5,7 ADH1 ARP4 ARP8 CAB3 CHD1 CKA1 CKA2 CKB1 CKB2 DPB2 DPB4 ESC8 HHF1, HHF2 HOT1 IES1 IES5 INO80 IOC3 ISW1 ISW2 ITC1 KAP114 MOT1 NAP1 NHP10 NIP1 NPL6 POL2 PRT1 PUF6 REB1 RET1 RFA1 RFX1 RNR2 RPA135 RPC34 RPC40 RPC82 RPG1 RPO31 RSC2 RSC3 RSC4 RSC58 RSC6 RSC8 RVB2 SFH1 SIN3 SPT16 SSA4 STH1 STO1 TAF1 TFC7 TIF35 TOP1 UTP22 VPS1 YBR090C-A, NHP6B YTA7 |
| View Details | Ho Y, et al. (2002) |
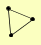
|
ISW1 ISW2 KAP95 |
| View Details | Qiu et al. (2008) |
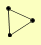
|
DLS1 ISW2 ITC1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
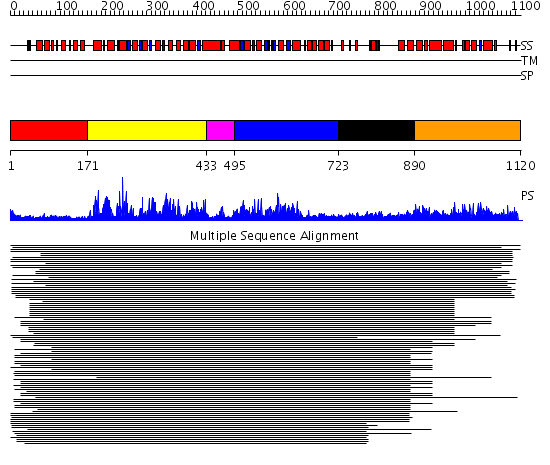
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..170] | 1.482995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [171..432] | 105.29243 | SNF2 family N-terminal domain No confident structure predictions are available. | |
| 3 | View Details | [433..494] | 4.522879 | Putative DEAD box RNA helicase | |
| 4 | View Details | [495..722] | 2.69897 | Nucleotide excision repair enzyme UvrB | |
| 5 | View Details | [723..889] | 2.211994 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [890..1120] | 1.066996 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)