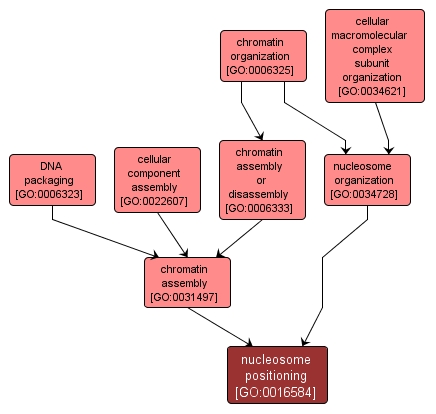
GO TERM SUMMARY
|
| Name: |
nucleosome positioning |
| Acc: |
GO:0016584 |
| Aspect: |
Biological Process |
| Desc: |
Ordering of successions of nucleosomes into regular arrays so that nucleosomes are positioned at defined distances from one another. |
| Synonyms:
|
|

|
INTERACTIVE GO GRAPH
|