













| General Information: |
|
| Name(s) found: |
YLR035C-A
[SGD]
|
| Description(s) found:
Found 15 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1155 amino acids |
Gene Ontology: |
|
| Cellular Component: |
retrotransposon nucleocapsid
[ISS |
| Biological Process: |
transposition, RNA-mediated
[ISS |
| Molecular Function: |
RNA binding
[ISS protein binding [ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLAHANAQTI RYSLKNNTIT YFNESDVDWS SAIDYQCPDC LIGKSTKHRH IKGSRLKYQN 60
61 SYEPFQYLHT DIFGPVHNLP KSAPSYFISF TDETTKFRWV YPLHDRREDS ILDVFTTILA 120
121 FIKNQFQASV LVIQMDRGSE YTNRTLHKFL EKNGITPCYT TTADSRAHGV AERLNRTLLD 180
181 DCRTQLQCSG LPNHLWFSAI EFSTIVRNSL ASPKSKKSAR QHAGLAGLDI STLLPFGQPV 240
241 IVNDHNPNSK IHPRGIPGYA LHPSRNSYGY IIYLPSLKKT VDTTNYVILQ GKESRLDQFN 300
301 YDALTFDEDL NRLTASYQSF IASNEIQQSD DLNIESDHDF QSDIELHPEQ PRNVLSKAVS 360
361 PTDSTPPSTH TEDSKRVSKT NIRAPREVDP NISESNILPS KKRSSTPQIS DIESTGSGGM 420
421 HRLDVPLLAP MSQSNTHESS HASKSKDFRH SDSYSDNETN HTNVPISSTG GTNNKTVPQT 480
481 SEQETEKRII HRSPSIDTSS SESNSLHHVV PIKTSDTCPK ENTEESIIAD LPLPDLPPEP 540
541 PTELSDSFKE LPPINSHQTN SSLGGIGDSN AYTTINSKKR SLEDNETEIK VSRDTWNTKN 600
601 MRSLEPPRSK KRIHLIAAVK AVKSIKPIRT TLRYDEAITY NKDIKEKEKY IQAYHKEVNQ 660
661 LLKMKTWDTD RYYDRKEIDP KRVINSMFIF NRKRGGTHKA RFVARGDIQH PDTYDPGMQS 720
721 NTVHHYALMT SLSLASDNNY YITQLDISSA YLYADIKEEL YIRPPPHLGM NDKLIRLKKS 780
781 LYGLKQSGAN WYETIKSYLI KQCGMEEVRG WSCVFKNSQV TICLFVDDMI LFSKDLNANK 840
841 KIITTLKKQY DTKIINLGES DNEIQYDILG LEIKYQRGKY MKLGMENSLT EKIPKLNVPL 900
901 NPKGRKLSAP GQPGLYIDQQ ELELEEDDYK MKVHEMQKLI GLASYVGYKF RFDLLYYINT 960
961 LAQHILFPSK QVLDMTYELI QFIWNTRDKQ LIWHKSKPVK PTNKLVVISD ASYGNQPYYK1020
1021 SQIGNIYLLN GKVIGGKSTK ASLTCTSTTE AEIHAISESV PLLNNLSHLV QELNKKPITK1080
1081 GLLTDSKSTI SIIISNNEEK FRNRFFGTKA MRLRDEVSGN HLHVCYIETK KNIADVMTKP1140
1141 LPIKTFKLLT NKWIH |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Hazbun TR, et al. (2003) |
|
AMN1 AXL2 IMH1 PEX19 PFK27 RAX2 RGT2 RIC1 RIX7 RPA190 SST2 TRX1 TRX2 YLR035C-A |
| View Details | Ho Y, et al. (2002) |

|
ADR1 BBC1 CDC39 CDC53 CRM1 DUR1,2 ECM29 ECM33 FAA4 GAL3 GCN1 HRT1 HYP2 MDN1 MKT1 MMS1 MYO2 PFK1 PMA2 RPA190 RPN1 TPS1 UBI4 VPS13 YAR009C YGP1 YLR035C-A |
| View Details | Qiu et al. (2008) |
|
YDR210C-C YDR210C-D YLR035C-A YOR343W-A YOR343W-B |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
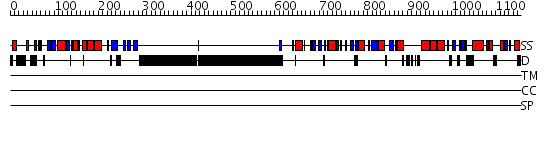
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..367] | 26.0 | N-terminal Zn binding domain of HIV integrase; Retroviral integrase, catalytic domain | |
| 2 | View Details | [368..629] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [630..1155] | 1.41 | HIV RNase H (Domain of reverse transcriptase); HIV-1 reverse transcriptase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)