













| General Information: |
|
| Name(s) found: |
RDS2 /
YPL133C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 446 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
response to xenobiotic stimulus
[IMP |
| Molecular Function: |
transcription factor activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSANSGVKRA SKAFKTCLFC KRSHVVCDKQ RPCSRCVKRD IAHLCREDDI AVPNEMPSQH 60
61 ESSPNDNNIQ GKYANKAHTG IPSDYQNEPV NKSGSTYGEE LSPKLDSSLV NDTTSLLLPQ 120
121 QPVFVSENVG SEFSSLNEFL SMLENPLLTQ TSLSSSSASN VHLENGSQTT QSPLEYQNDN 180
181 RRDEIGVARQ ENRSPTIMSG SSNSISKGDK QDQEKEESRI LANANENSAP TPKEQFFLTA 240
241 ADPSTEMTPE HRLKLVINAK LEAGLLKPYN YAKGYARLQD YMDKYMNQSS KQRILKPLST 300
301 IRPAFRTIAR SLKDVDLVLV EESFERMLLS YDRVFTSMSM PACLCRRTGE IYRANKEFAS 360
361 LVDCTVDDLR DGKLAIYELM TEESAVNFWE KYGSIAFDKG QKAVLTSCSL RTKDGIRKRP 420
421 CCFSFTIRRD RYNIPICIVG NFIPLS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
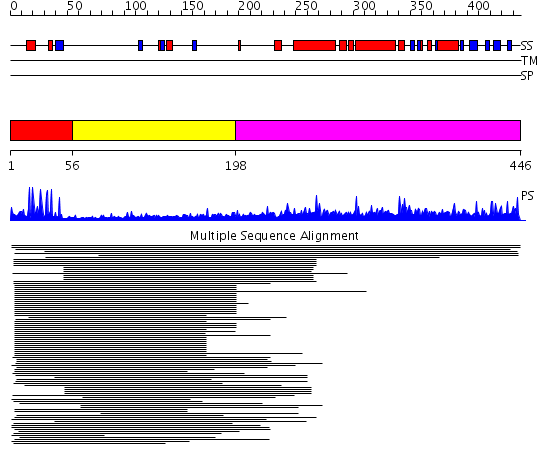
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..55] | 30.457575 | Hap1 (Cyp1) | |
| 2 | View Details | [56..197] | 3.276997 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [198..446] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [269..446] | 1.77 | Photoactive yellow protein, PYP |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)