













| General Information: |
|
| Name(s) found: |
PEX1 /
YKL197C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1043 amino acids |
Gene Ontology: |
|
| Cellular Component: |
peroxisome
[IDA |
| Biological Process: |
peroxisome organization
[TAS]
|
| Molecular Function: |
ATPase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTTKRLKFE NLRIQFSNAI VGNFLRLPHS IINVLESTNY AIQEFGIAVH SHNSDIPIVH 60
61 LGWDGHDSGS SENVVLINPV LATVYDLNQK SPLVDLYIQR YDHTHLATEV YVTPETSDDW 120
121 EIIDANAMRF QNGEILHQTR IVTPGETLIC YLEGIVTKFK IDRVEPSMKS ARITDGSLVV 180
181 VAPKVNKTRL VKAEYGHSNK TILKNGAIQL LKKVILRSTV CKMDFPKDNL FVVYISDGAQ 240
241 LPSQKGYASI VKCSLRQSKK SDSDNKSVGI PSKKIGVFIK CDSQIPENHI ALSSHLWDAF 300
301 FTHPMNGAKI KLEFLQMNQA NIISGRNATV NIKYFGKDVP TKSGDQYSKL LGGSLLTNNL 360
361 ILPTEQIIIE IKKGESEQQL CNLNEISNES VQWKVTQMGK EEVKDIIERH LPKHYHVKET 420
421 GEVSRTSKDE DDFITVNSIK KEMVNYLTSP IIATPAIILD GKQGIGKTRL LKELINEVEK 480
481 DHHIFVKYAD CETLHETSNL DKTQKLIMEW CSFCYWYGPS LIVLDNVEAL FGKPQANDGD 540
541 PSNNGQWDNA SKLLNFFINQ VTKIFNKDNK RIRVLFSGKQ KTQINPLLFD KHFVSETWSL 600
601 RAPDKHARAK LLEYFFSKNQ IMKLNRDLQF SDLSLETEGF SPLDLEIFTE KIFYDLQLER 660
661 DCDNVVTREL FSKSLSAFTP SALRGVKLTK ETNIKWGDIG ALANAKDVLL ETLEWPTKYE 720
721 PIFVNCPLRL RSGILLYGYP GCGKTLLASA VAQQCGLNFI SVKGPEILNK FIGASEQNIR 780
781 ELFERAQSVK PCILFFDEFD SIAPKRGHDS TGVTDRVVNQ LLTQMDGAEG LDGVYILAAT 840
841 SRPDLIDSAL LRPGRLDKSV ICNIPTESER LDILQAIVNS KDKDTGQKKF ALEKNADLKL 900
901 IAEKTAGFSG ADLQGLCYNA YLKSVHRWLS AADQSEVVPG NDNIEYFSIN EHGRREENRL 960
961 RLKTLLQQDV VHETKTSTSA ASELTAVVTI NDLLEACQET KPSISTSELV KLRGIYDRFQ1020
1021 KDRNGEMPNG ENSIDIGSRL SLM |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
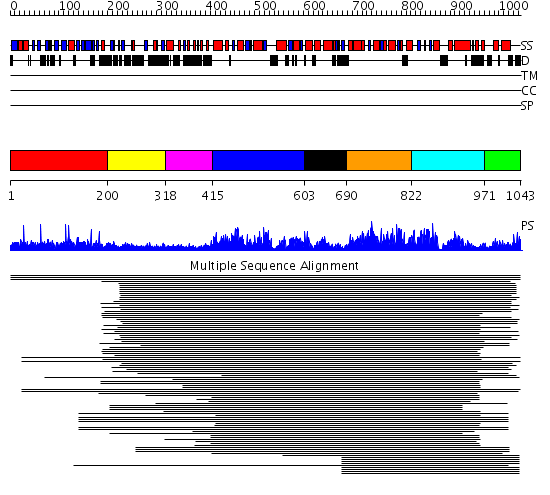
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 7.29 | N-terminal domain of VAT-N, VAT-Nn; C-terminal domain of VAT-N, VAT-Nc | |
| 2 | View Details | [200..317] | 526.457575 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 3 | View Details | [318..414] | 526.457575 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 4 | View Details | [415..602] | 526.457575 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 5 | View Details | [603..689] | 526.457575 | Membrane fusion atpase p97 N-terminal domain , P97-Nn; Membrane fusion atpase p97, D1 domain; Membrane fusion atpase p97 domain 2, P97-Nc | |
| 6 | View Details | [690..821] | 5.69897 | HslU | |
| 7 | View Details | [822..970] | 13.02 | Holliday junction helicase RuvB | |
| 8 | View Details | [971..1043] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)