













| General Information: |
|
| Name(s) found: |
OTU1 /
YFL044C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 301 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
protein deubiquitination
[IDA ubiquitin cycle [IGI regulation of transcription [IMP |
| Molecular Function: |
ubiquitin-specific protease activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKLKVTGAGI NQVVTLKQDA TLNDLIEHIN VDVKTMRFGY PPQRINLQGE DASLGQTQLD 60
61 ELGINSGEKI TIESSDSNES FSLPPPQPKP KRVLKSTEMS IGGSGENVLS VHPVLDDNSC 120
121 LFHAIAYGIF KQDSVRDLRE MVSKEVLNNP VKFNDAILDK PNKDYAQWIL KMESWGGAIE 180
181 IGIISDALAV AIYVVDIDAV KIEKFNEDKF DNYILILFNG IHYDSLTMNE FKTVFNKNQP 240
241 ESDDVLTAAL QLASNLKQTG YSFNTHKAQI KCNTCQMTFV GEREVARHAE STGHVDFGQN 300
301 R |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
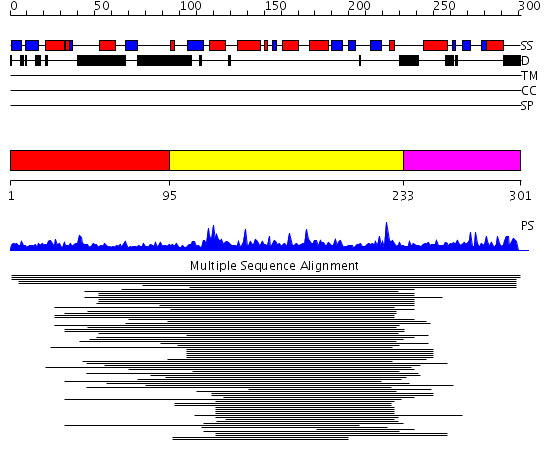
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..94] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [95..232] | 38.79588 | OTU-like cysteine protease No confident structure predictions are available. | |
| 3 | View Details | [233..301] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)