













| General Information: |
|
| Name(s) found: |
POL3 /
YDL102W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1097 amino acids |
Gene Ontology: |
|
| Cellular Component: |
replication fork
[IDA delta DNA polymerase complex [TAS |
| Biological Process: |
mutagenesis
[TAS lagging strand elongation [TAS base-excision repair [TAS leading strand elongation [TAS maintenance of fidelity involved in DNA-dependent DNA replication [IGI nucleotide-excision repair [TAS mismatch repair [NAS postreplication repair [TAS |
| Molecular Function: |
single-stranded DNA specific 3'-5' exodeoxyribonuclease activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEKRSLPMV DVKIDDEDTP QLEKKIKRQS IDHGVGSEPV STIEIIPSDS FRKYNSQGFK 60
61 AKDTDLMGTQ LESTFEQDVS QMEHDMADQE EHDLSSFERK KLPTDFDPSL YDISFQQIDA 120
121 EQSVLNGIKD ENTSTVVRFF GVTSEGHSVL CNVTGFKNYL YVPAPNSSDA NDQEQINKFV 180
181 HYLNETFDHA IDSIEVVSKQ SIWGYSGDTK LPFWKIYVTY PHMVNKLRTA FERGHLSFNS 240
241 WFSNGTTTYD NIAYTLRLMV DCGIVGMSWI TLPKGKYSMI EPNNRVSSCQ LEVSINYRNL 300
301 IAHPAEGDWS HTAPLRIMSF DIECAGRIGV FPEPEYDPVI QIANVVSIAG AKKPFIRNVF 360
361 TLNTCSPITG SMIFSHATEE EMLSNWRNFI IKVDPDVIIG YNTTNFDIPY LLNRAKALKV 420
421 NDFPYFGRLK TVKQEIKESV FSSKAYGTRE TKNVNIDGRL QLDLLQFIQR EYKLRSYTLN 480
481 AVSAHFLGEQ KEDVHYSIIS DLQNGDSETR RRLAVYCLKD AYLPLRLMEK LMALVNYTEM 540
541 ARVTGVPFSY LLARGQQIKV VSQLFRKCLE IDTVIPNMQS QASDDQYEGA TVIEPIRGYY 600
601 DVPIATLDFN SLYPSIMMAH NLCYTTLCNK ATVERLNLKI DEDYVITPNG DYFVTTKRRR 660
661 GILPIILDEL ISARKRAKKD LRDEKDPFKR DVLNGRQLAL KISANSVYGF TGATVGKLPC 720
721 LAISSSVTAY GRTMILKTKT AVQEKYCIKN GYKHDAVVVY GDTDSVMVKF GTTDLKEAMD 780
781 LGTEAAKYVS TLFKHPINLE FEKAYFPYLL INKKRYAGLF WTNPDKFDKL DQKGLASVRR 840
841 DSCSLVSIVM NKVLKKILIE RNVDGALAFV RETINDILHN RVDISKLIIS KTLAPNYTNP 900
901 QPHAVLAERM KRREGVGPNV GDRVDYVIIG GNDKLYNRAE DPLFVLENNI QVDSRYYLTN 960
961 QLQNPIISIV APIIGDKQAN GMFVVKSIKI NTGSQKGGLM SFIKKVEACK SCKGPLRKGE1020
1021 GPLCSNCLAR SGELYIKALY DVRDLEEKYS RLWTQCQRCA GNLHSEVLCS NKNCDIFYMR1080
1081 VKVKKELQEK VEQLSKW |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
POL3 POL31 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
CDC45 DPB11 DPB2 DPB3 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC1 ORC2 ORC3 ORC4 ORC5 ORC6 POL2 POL3 POL31 POL32 |
| View Details | Krogan NJ, et al. (2006) |
|
POL3 POL30 POL31 POL32 |
| View Details | Gavin AC, et al. (2006) |

|
POL3 POL31 |
| View Details | Qiu et al. (2008) |
|
CDC45 CDC9 DPB11 DPB2 DPB3 DPB4 HCS1 MCM2 MCM3 MCM4 MCM5 MCM6 MCM7 ORC2 ORC3 ORC5 POL1 POL12 POL2 POL3 POL30 POL31 POL32 PRI1 PRI2 RAD30 REV3 REV7 RFC1 RFC4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample bob1 (2nd set) from october 2005 | McCusker D, et al (2007) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
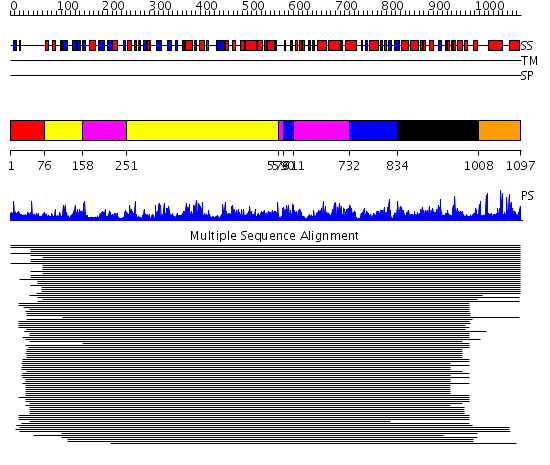
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [76..157] [251..578] |
1532.218487 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 3 | View Details | [158..250] [579..589] [611..731] |
1532.218487 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 4 | View Details | [590..610] [732..833] |
1532.218487 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 5 | View Details | [834..1007] | 1532.218487 | Exonuclease domain of family B (archaeal and phage) DNA polymerases; T4-like DNA polymerase | |
| 6 | View Details | [1008..1097] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)