













| General Information: |
|
| Name(s) found: |
IRA1 /
YBR140C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 3092 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA membrane [IMP |
| Biological Process: |
negative regulation of Ras protein signal transduction
[IMP regulation of adenylate cyclase activity [IMP negative regulation of cAMP biosynthetic process [IMP positive regulation of Ras GTPase activity [IMP |
| Molecular Function: |
Ras GTPase activator activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNQSDPQDKK NFPMEYSLTK HLFFDRLLLV LPIESNLKTY ADVEADSVFN SCRSIILNIA 60
61 ITKDLNPIIE NTLGLIDLIV QDEEITSDNI TDDIAHSILV LLRLLSDVFE YYWDQNNDFK 120
121 KIRNDNYKPG FSSHRPNFHT SRPKHTRINP ALATMLLCKI SKLKFNTRTL KVLQNMSHHL 180
181 SGSATISKSS ILPDSQEFLQ KRNYPAYTEK IDLTIDYIQR FISASNHVEF TKCVKTKVVA 240
241 PLLISHTSTE LGVVNHLDLF GCEYLTDKNL LAYLDILQHL SSYMKRTIFH SLLLYYASKA 300
301 FLFWIMARPK EYVKIYNNLI SSDYNSPSSS SDNGGSNNSD KTSISQLVSL LFDDVYSTFS 360
361 VSSLLTNVNN DHHYHLHHSS SSSKTTNTNS PNSISKTSIK QSSVNASGNV SPSQFSTGND 420
421 ASPTSPMASL SSPLNTNILG YPLSPITSTL GQANTSTSTT AATTKTDADT PSTMNTNNNN 480
481 NNNNSANLNN IPQRIFSLDD ISSFNSSRKS LNLDDSNSLF LWDTSQHSNA SMTNTNMHAG 540
541 VNNSQSQNDQ SSLNYMENIM ELYSNYTGSE LSSHTAILRF LVVLTLLDSE VYDEMNSNSY 600
601 RKISEPIMNI NPKDSNTSSW GSASKNPSIR HLTHGLKKLT LQQGRKRNVK FLTYLIRNLN 660
661 GGQFVSDVSL IDSIRSILFL MTMTSSISQI DSNIASVIFS KRFYNLLGQN LEVGTNWNSA 720
721 TANTFISHCV ERNPLTHRRL QLEFFASGLQ LDSDLFLRHL QLEKELNHID LPKISLYTEG 780
781 FRVFFHLVST KKLHEDIAEK TSSVLKRLFC IIADILLKAT PYFDDNVTKI IASILDGHIL 840
841 DQFDAARTLS NDDHVSFDAA TSVYTEPTEI IHNSSDASLV SSLSQSPLSI NSGSNITNTR 900
901 TWDIQSILPT LSNRSSASDL SLSNILTNPL EAQQNNNANL LAHRLSGVPT TKRYASPNDS 960
961 ERSRQSPYSS PPQLQQSDLP SPLSVLSSSA GFSSNHSITA TPTILKNIKS PKPNKTKKIA1020
1021 DDKQLKQPSY SRVILSDNDE ARKIMMNIFS IFKRMTNWFI RPDANTEFPK TFTDIIKPLF1080
1081 VSILDSNQRL QVTARAFIEI PLSYIATFED IDNDLDPRVL NDHYLLCTYA VTLFASSLFD1140
1141 LKLENAKREM LLDIIVKFQR VRSYLSNLAE KHNLVQAIIT TERLTLPLLV GAVGSGIFIS1200
1201 LYCSRGNTPR LIKISCCEFL RSLRFYQKYV GALDQYSIYN IDFIDAMAQD NFTASGSVAL1260
1261 QRRLRNNILT YIKGSDSILL DSMDVIYKKW FYFSCSKSVT QEELVDFRSL AGILASMSGI1320
1321 LSDMQELEKS KSAPDNEGDS LSFESRNPAY EVHKSLKLEL TKKMNFFISK QCQWLNNPNL1380
1381 LTRENSRDIL SIELHPLSFN LLFNNLGLKI DELMSIDLSK SHEDSSFVLL EQIIIIIRTI1440
1441 LKRDDDEKIM LLFSTDLLDA VDKLIEIVEK ISIKSSKYYK GIIQMSKMFR AFEHSEKNLG1500
1501 ISNHFHLKNK WLKLVIGWFK LSINKDYDFE NLSRPLREMD LQKRDEDFLY IDTSIESAKA1560
1561 LAYLTHNVPL EIPPSSSKED WNRSSTVSFG NHFTILLKGL EKSADLNQFP VSLRHKISIL1620
1621 NENVIIALTN LSNANVNVSL KFTLPMGYSP NKDIRIAFLR VFIDIVTNYP VNPEKHEMDK1680
1681 MLAIDDFLKY IIKNPILAFF GSLACSPADV DLYAGGFLNA FDTRNASHIL VTELLKQEIK1740
1741 RAARSDDILR RNSCATRALS LYTRSRGNKY LIKTLRPVLQ GIVDNKESFE IDKMKPGSEN1800
1801 SEKMLDLFEK YMTRLIDAIT SSIDDFPIEL VDICKTIYNA ASVNFPEYAY IAVGSFVFLR1860
1861 FIGPALVSPD SENIIIVTHA HDRKPFITLA KVIQSLANGR ENIFKKDILV SKEEFLKTCS1920
1921 DKIFNFLSEL CKIPTNNFTV NVREDPTPIS FDYSFLHKFF YLNEFTIRKE IINESKLPGE1980
1981 FSFLKNTVML NDKILGVLGQ PSMEIKNEIP PFVVENREKY PSLYEFMSRY AFKKVDMKEE2040
2041 EEDNAPFVHE AMTLDGIQII VVTFTNCEYN NFVMDSLVYK VLQIYARMWC SKHYVVIDCT2100
2101 TFYGGKANFQ KLTTLFFSLI PEQASSNCMG CYYFNVNKSF MDQWASSYTV ENPYLVTTIP2160
2161 RCFINSNTDQ SLIKSLGLSG RSLEVLKDVR VTLHDITLYD KEKKKFCPVS LKIGNKYFQV2220
2221 LHEIPQLYKV TVSNRTFSIK FNNVYKISNL ISVDVSNTTG VSSEFTLSLD NEEKLVFCSP2280
2281 KYLEIVKMFY YAQLKMEEDF GTDFSNDISF STSSSAVNAS YCNVKEVGEI ISHLSLVILV2340
2341 GLFNEDDLVK NISYNLLVAT QEAFNLDFGT RLHKSPETYV PDDTTTFLAL IFKAFSESST2400
2401 ELTPYIWKYM LDGLENDVIP QEHIPTVVCS LSYWVPNLYE HVYLANDEEG PEAISRIIYS2460
2461 LIRLTVKEPN FTTAYLQQIW FLLALDGRLT NVIVEEIVSH ALDRDSENRD WMKAVSILTS2520
2521 FPTTEIACQV IEKLINMIKS FLPSLAVEAS AHSWSELTIL SKISVSIFFE SPLLSQMYLP2580
2581 EILFAVSLLI DVGPSEIRVS LYELLMNVCH SLTNNESLPE RNRKNLDIVC ATFARQKLNF2640
2641 ISGFSQEKGR VLPNFAASSF SSKFGTLDLF TKNIMLLMEY GSISEGAQWE AKYKKYLMDA2700
2701 IFGHRSFFSA RAMMILGIMS KSHTSLFLCK ELLVETMKVF AEPVVDDEQM FIIIAHVFTY2760
2761 SKIVEGLDPS SELMKELFWL ATICVESPHP LLFEGGLLFM VNCLKRLYTV HLQLGFDGKS2820
2821 LAKKLMESRN FAATLLAKLE SYNGCIWNED NFPHIILGFI ANGLSIPVVK GAALDCLQAL2880
2881 FKNTYYERKS NPKSSDYLCY LFLLHLVLSP EQLSTLLLEV GFEDELVPLN NTLKVPLTLI2940
2941 NWLSSDSDKS NIVLYQGALL FSCVMSDEPC KFRFALLMRY LLKVNPICVF RFYTLTRKEF3000
3001 RRLSTLEQSS EAVAVSFELI GMLVTHSEFN YLEEFNDEMV ELLKKRGLSV VKPLDIFDQE3060
3061 HIEKLKGEGE HQVAIYERKR LATMILARMS CS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
IRA1 RPL17B |
| View Details | Krogan NJ, et al. (2006) |
|
AIM26 APC9 BEM3 FMP27 IRA1 MDM36 MID1 NUP42 REV1 SEC20 SML1 TCO89 TOP1 YDR396W YGL024W ZAP1 ZIM17 |
| View Details | Ho Y, et al. (2002) |
|
CDC25 CKI1 GCR2 GIN4 HOM3 IRA1 IRA2 MYO2 MYO4 NAP1 PMD1 PRS2 PRS3 PRS5 RAS2 RIM11 TPS1 TSL1 TSR1 YDR170W-A YER138C YER160C YJR027W YJR028W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
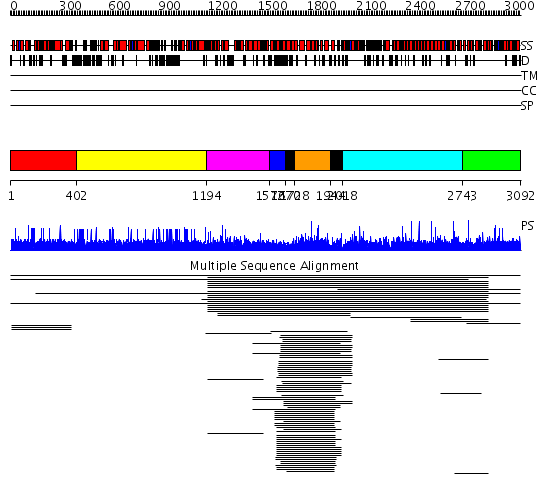
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..401] | 1.004848 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [402..1193] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [1194..1571] | 3.016942 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [1572..1669] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [1670..1727] [1944..2017] |
280.68867 | GAP related domain of neurofibromin | |
| 6 | View Details | [1728..1943] | 280.68867 | GAP related domain of neurofibromin | |
| 7 | View Details | [2018..2742] | 1.019001 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [2743..3092] | 1.012845 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)