













| Protein: | SPE3 |
| From Publication: | Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002 Jan 10;415(6868):180-3. |
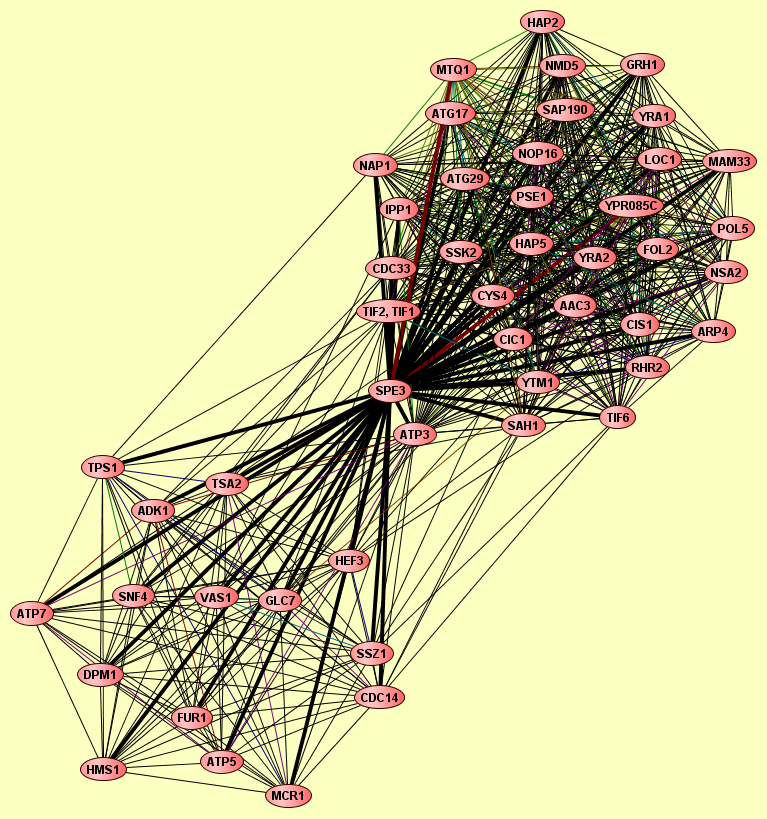
| Complexes containing SPE3: | 2 |
| LEGEND: |
|||
| = Same process, or one is unknown. | = Same branch, distance 4. | ||
| = Same branch, distance 1. | = Same branch, distance 5. | ||
| = Same branch, distance 2. | = Same branch, distance 6 or more. | ||
| = Same branch, distance 3. | = Not in same branch of GO. | ||
| Size | Notes | Members | |
| View GO Analysis | 17 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Cdc14. The topolgies of protein complexes in this experiment are unknown. | ADK1 ATP3 ATP5 ATP7 CDC14 DPM1 FUR1 GLC7 HEF3 HMS1 MCR1 SNF4 SPE3 SSZ1 TPS1 TSA2 VAS1 |
| View GO Analysis | 34 proteins | This molecular complex record represents a population of complexes co-purified by immunoprecipitation of FLAG-tagged Hap2. The topolgies of protein complexes in this experiment are unknown. | AAC3 ARP4 ASA1 ATG17 ATG29 ATP3 CDC33 CIC1 CIS1 CYS4 FOL2 GRH1 HAP2 HAP5 IPP1 LOC1 MAM33 MTQ1 NAP1 NMD5 NOP16 NSA2 POL5 PSE1 RHR2 SAH1 SAP190 SPE3 SSK2 TIF1, TIF2 TIF6 YRA1 YRA2 YTM1 |