













| General Information: |
|
| Name(s) found: |
lit-1 /
CE37161
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 454 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
embryonic development
[IMP embryonic cleavage [IMP polarity specification of proximal/distal axis [IMP spindle organization [IMP hermaphrodite genitalia development [IMP signal transduction [IEA embryonic development ending in birth or egg hatching [IMP asymmetric cell division [IMP protein amino acid phosphorylation [IEA reproduction [IMP asymmetric protein localization involved in cell fate determination [IMP nematode larval development [IMP Wnt receptor signaling pathway [IGI |
| Molecular Function: |
ATP binding
[IEA protein tyrosine kinase activity [IEA non-membrane spanning protein tyrosine kinase activity [IEA MAP kinase activity [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA kinase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MALVSHTHPA AVGSTTCYEK NQQKQQQVQQ IPTQPQVAHV SSNAILAAAQ PFYPPPVQDS 60
61 QPDRPIGYGA FGVVWSVTDP RSGKRVALKK MPNVFQNLAS CKRVFREIKM LSSFRHDNVL 120
121 SLLDILQPAN PSFFQELYVL TELMQSDLHK IIVSPQALTP DHVKVFVYQI LRGLKYLHTA 180
181 NILHRDIKPG NLLVNSNCIL KICDFGLART WDQRDRLNMT HEVVTQYYRA PELLMGARRY 240
241 TGAVDIWSVG CIFAELLQRK ILFQAAGPIE QLQMIIDLLG TPSQEAMKYA CEGAKNHVLR 300
301 AGLRAPDTQR LYKIASPDDK NHEAVDLLQK LLHFDPDKRI SVEEALQHRY LEEGRLRFHS 360
361 CMCSCCYTKP NMPSRLFAQD LDPRHESPFD PKWEKDMSRL SMFELREKMY QFVMDRPALY 420
421 GVALCINPQS AAYKNFASSS VAQASELPPS PQAW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
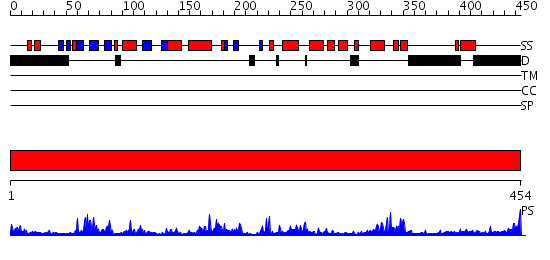
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..454] | 83.522879 | MAP kinase p38 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)