













| General Information: |
|
| Name(s) found: |
lit-1 /
CE29477
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 462 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA |
| Biological Process: |
embryonic development
[IMP embryonic cleavage [IMP polarity specification of proximal/distal axis [IMP spindle organization [IMP hermaphrodite genitalia development [IMP signal transduction [IEA embryonic development ending in birth or egg hatching [IMP asymmetric cell division [IMP protein amino acid phosphorylation [IEA reproduction [IMP asymmetric protein localization involved in cell fate determination [IMP nematode larval development [IMP Wnt receptor signaling pathway [IGI |
| Molecular Function: |
ATP binding
[IEA protein tyrosine kinase activity [IEA non-membrane spanning protein tyrosine kinase activity [IEA MAP kinase activity [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA kinase activity [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRDMIYGEMA LVSHTHPAAV GSTTCYEKNQ QKQQQVQQIP TQPQVAHVSS NAILAAAQPF 60
61 YPPPVQDSQP DRPIGYGAFG VVWSVTDPRS GKRVALKKMP NVFQNLASCK RVFREIKMLS 120
121 SFRHDNVLSL LDILQPANPS FFQELYVLTE LMQSDLHKII VSPQALTPDH VKVFVYQILR 180
181 GLKYLHTANI LHRDIKPGNL LVNSNCILKI CDFGLARTWD QRDRLNMTHE VVTQYYRAPE 240
241 LLMGARRYTG AVDIWSVGCI FAELLQRKIL FQAAGPIEQL QMIIDLLGTP SQEAMKYACE 300
301 GAKNHVLRAG LRAPDTQRLY KIASPDDKNH EAVDLLQKLL HFDPDKRISV EEALQHRYLE 360
361 EGRLRFHSCM CSCCYTKPNM PSRLFAQDLD PRHESPFDPK WEKDMSRLSM FELREKMYQF 420
421 VMDRPALYGV ALCINPQSAA YKNFASSSVA QASELPPSPQ AW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
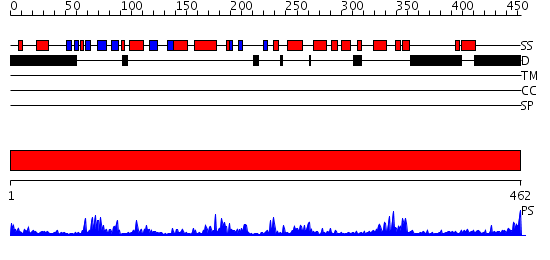
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..462] | 86.69897 | No description for 2gtmA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)