













| General Information: |
|
| Name(s) found: |
sty1 /
SPAC24B11.06c
[Sanger Pombe]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 349 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA cytosol [IDA |
| Biological Process: |
regulation of cAMP-mediated signaling
[IMP regulation of histone acetylation [IMP cellular sodium ion homeostasis [IGI protein amino acid phosphorylation [IGI regulation of meiotic cell cycle [IMP regulation of DNA binding [IDA cellular response to heat [TAS regulation of translation in response to nitrogen starvation [IDA response to arsenic [IMP cellular response to UV [TAS mRNA export from nucleus in response to heat stress [IMP cellular cation homeostasis [IGI stress-activated MAPK cascade [TAS response to osmotic stress [TAS cellular response to starvation [TAS response to cation stress [IGI response to hydrogen peroxide [IMP regulation of translation in response to oxidative stress [IDA positive regulation of mitotic cell cycle [IGI cellular response to nitrogen starvation [IMP regulation of translation in response to osmotic stress [IDA response to methylglyoxal [IDA regulation of chromatin disassembly [IMP regulation of chromatin assembly [IMP cellular response to oxidative stress [TAS cellular protein localization [IGI regulation of reciprocal meiotic recombination [IMP |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI MAP kinase activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEFIRTQIF GTCFEITTRY SDLQPIGMGA FGLVCSAKDQ LTGMNVAVKK IMKPFSTPVL 60
61 AKRTYRELKL LKHLRHENII SLSDIFISPF EDIYFVTELL GTDLHRLLTS RPLETQFIQY 120
121 FLYQILRGLK FVHSAGVIHR DLKPSNILIN ENCDLKICDF GLARIQDPQM TGYVSTRYYR 180
181 APEIMLTWQK YNVEVDIWSA GCIFAEMIEG KPLFPGRDHV NQFSIITELL GTPPMEVIET 240
241 ICSKNTLRFV QSLPQKEKVP FAEKFKNADP DAIDLLEKML VFDPRKRISA ADALAHNYLA 300
301 PYHDPTDEPV ADEVFDWSFQ DNDLPVETWK VMMYSEVLSF HNMDNELQS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
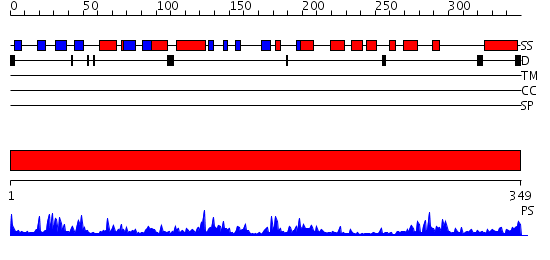
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..349] | 95.154902 | MAP kinase p38 |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)