













| General Information: |
|
| Name(s) found: |
MSH1 /
YHR120W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 959 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrion
[IDA |
| Biological Process: |
DNA repair
[IMP [NOT]reciprocal meiotic recombination [IMP mismatch repair [IMP mitochondrion organization [IMP mitochondrial genome maintenance [IMP |
| Molecular Function: |
DNA-dependent ATPase activity
[IDA dinucleotide insertion or deletion binding [IDA guanine/thymine mispair binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKHFFRLPTA FRPISRVSLR YSSTDTAQPK ISKLKISFNK ISESNSEKKD NLGSIDTRNC 60
61 LSTQQDDKLS STEPSKASLP PSLQYVRDLM DLYKDHVVLT QMGSFYELYF EQAIRYAPEL 120
121 NISLTNRAYS HGKVPFAGFP VHQLSRHLKM LVNNCGYSVT IAEQFKKKDV ADNEANKFYR 180
181 RVTRIVTPGT FIDEAFENLR ENTYLLNIEF PENCMSQVAD TSLKVGICWC DVSTGEIFVQ 240
241 QVYLRDLVSA ITRIQPKEIL LDERLLEFHI ESGTWYPELV ELKKFFIKYQ KMPSQHRTIE 300
301 SFYGLFNLGG KEATERQLKI QFQTFTQKEL AALRNTLIYV SNHLPDFSIN FQIPQRQLAT 360
361 AIMQIDSRTS TALELHSTVR DNNKKGSLLS SIRRTVTPSG TRLLSQWLSG PSLDLKEIKK 420
421 RQKIVAFFKD NRDITETLRT MLKKVNDLSR ILQKFSFGRG EALELIQMAR SLEVSREIRK 480
481 YLLNNTSLMK ATLKSQITQL TESLNFEKNL IDDILKFLNE EELAKSQDAK QNADVTRMLD 540
541 IDVKDKKESN KDEIFELRDF IVNPSFNTKL RKLHDTYQGV WQKKTEYNAL LKGFFVGDLG 600
601 AKTFTLKERQ NGEYALHVTG TASSLKKIDE LISKSTEYHG SCFHILQKSS QTRWLSHKIW 660
661 TDLGHELELL NLKIRNEEAN IIDLFKRKFI DRSNEVRQVA TTLGYLDTLS SFAVLANERN 720
721 LVCPKVDESN KLEVVNGRHL MVEEGLSARS LETFTANNCE LAKDNLWVIT GPNMGGKSTF 780
781 LRQNAIIVIL AQIGCFVPCS KARVGIVDKL FSRVGSADDL YNEMSTFMVE MIETSFILQG 840
841 ATERSLAILD EIGRGTSGKE GISIAYATLK YLLENNQCRT LFATHFGQEL KQIIDNKCSK 900
901 GMSEKVKFYQ SGITDLGGNN FCYNHKLKPG ICTKSDAIRV AELAGFPMEA LKEAREILG |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
MAS1 MAS2 MSH1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #30 Asynchronous Prep6-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #17 Mitotic Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
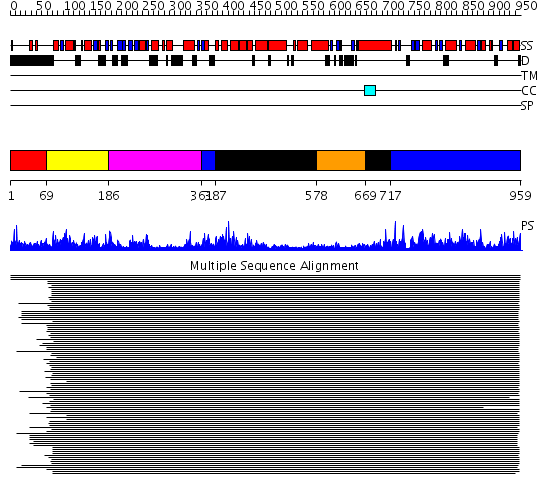
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [69..185] | 1421.54902 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 3 | View Details | [186..360] | 1421.54902 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 4 | View Details | [361..386] [717..959] |
1421.54902 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 5 | View Details | [387..577] [669..716] |
1421.54902 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I | |
| 6 | View Details | [578..668] | 1421.54902 | DNA repair protein MutS, domain III; DNA repair protein MutS, the C-terminal domain; DNA repair protein MutS, domain II; DNA repair protein MutS, domain I |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)