













| General Information: |
|
| Name(s) found: |
SRP101 /
YDR292C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 621 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum membrane
[IDA signal recognition particle receptor complex [TAS |
| Biological Process: |
SRP-dependent cotranslational protein targeting to membrane
[IC protein targeting to ER [IMP |
| Molecular Function: |
GTP binding
[IPI signal recognition particle binding [IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFDQLAVFTP QGQVLYQYNC LGKKFSEIQI NSFISQLITS PVTRKESVAN ANTDGFDFNL 60
61 LTINSEHKNS PSFNALFYLN KQPELYFVVT FAEQTLELNQ ETQQTLALVL KLWNSLHLSE 120
121 SILKNRQGQN EKNKHNYVDI LQGIEDDLKK FEQYFRIKYE ESIKQDHINP DNFTKNGSVP 180
181 QSHNKNTKKK LRDTKGKKQS TGNVGSGRKW GRDGGMLDEM NHEDAAKLDF SSSNSHNSSQ 240
241 VALDSTINKD SFGDRTEGGD FLIKEIDDLL SSHKDEITSG NEAKNSGYVS TAFGFLQKHV 300
301 LGNKTINESD LKSVLEKLTQ QLITKNVAPE AADYLTQQVS HDLVGSKTAN WTSVENTARE 360
361 SLTKALTQIL TPGVSVDLLR EIQSKRSKKD EEGKCDPYVF SIVGVNGVGK STNLSKLAFW 420
421 LLQNNFKVLI VACDTFRSGA VEQLRVHVEN LAQLMDDSHV RGSKNKRGKT GNDYVELFEA 480
481 GYGGSDLVTK IAKQAIKYSR DQNFDIVLMD TAGRRHNDPT LMSPLKSFAD QAKPDKIIMV 540
541 GEALVGTDSV QQAKNFNDAF GKGRNLDFFI ISKCDTVGEM LGTMVNMVYA TGIPILFVGV 600
601 GQTYTDLRTL SVKWAVNTLM S |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
SRP101 SRP102 |
| View Details | (MIPS) Mewes HW, et al. (2004) |

|
SRP101 SRP102 |
| View Details | Gavin AC, et al. (2006) |

|
SRP101 SRP102 |
| View Details | Gavin AC, et al. (2002) |

|
SRP101 SRP102 |
| View Details | Qiu et al. (2008) |
|
SBH1 SRP101 SRP102 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
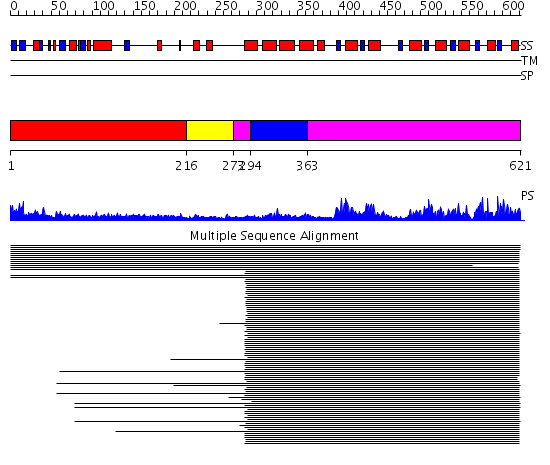
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..215] | 4.209996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [216..272] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [273..293] [363..621] |
271.491979 | Signal recognition particle receptor, FtsY; GTPase domain of the signal recognition particle receptor FtsY | |
| 4 | View Details | [294..362] | 271.491979 | Signal recognition particle receptor, FtsY; GTPase domain of the signal recognition particle receptor FtsY |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)