













| General Information: |
|
| Name(s) found: |
ESP1 /
YGR098C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1630 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA mitochondrion [IDA spindle [IDA |
| Biological Process: |
regulation of exit from mitosis
[IGI mitotic sister chromatid segregation [IMP |
| Molecular Function: |
cysteine-type endopeptidase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMVKQEEPLN EISPNTPMTS KSYLLNDTLS KVHHSGQTRP LTSVLSGDAS SNSIGILAMH 60
61 NNIIRDFTKI ASNNIDLAIE DITTVDHSLN SIYSLLKSHH MWGHINSTVK QHLMIIVKLI 120
121 NNNALGLASS EIIFLFNETN LFQAHSLKNI LLADFSTWND YYLSNLKILA LQIILKRKLV 180
181 DEYLPHILEL FSHDKRYLLK DPNLKAHALT KIVLSFFSVT TSCKVLFGLK FLQYIKQFKL 240
241 PFKKFISNIT VECFSKNLLH KNYLEMGPNK IYLNSFYLSY SMLYDGLDKI MLLDILSYEE 300
301 TTEVQRAIKS KKEFNEYCNM SENRLLWSCI SVDDLNVILE NATNFLQNKG KHISATLKCL 360
361 VCLWSTIRLE GLPKNKDILR QFDCTVIYIN SNIKSINDES AAALLSELLG VLSEICIDYK 420
421 EPKRLSNIIS VLFNASVLFK SHSFLLKTAN LEISNVLISN DSKTSHRTIL KFEKFISSAQ 480
481 SAQKKIEIFS CLFNVYCMLR NDTLSFVFDF CQNAFIHCFT RLKITKFIEF SNSSEIMLSV 540
541 LYGNSSIENI PSENWSQLSR MIFCSLRGIF DLDPLELNNT FDKLHLLNKY ELLIRIVYLL 600
601 NLDMSKHLTT NLSKITKLYI NKWLQKSDEK AERISSFEMD FVKMLLCYLN FNNFDKLSIE 660
661 LSLCIKSKEK YYSSIVPYAD NYLLEAYLSL YMIDDALMMK NQLQKTMNLS TAKIEQALLH 720
721 ASSLINVHLW DSDLTAFQIY FGKTLPAMKP ELFDINNDHN LPMSLYIKVI LLNIKIFNES 780
781 AKLNIKAGNV ISAVIDCRKA QNLALSLLKK KNKLSQGSRL ALLKSLSFSF FQLIKIHIRI 840
841 GSARDCEFYS KELSRIISDL EEPIIVYRCL HFLHRYYMIT EQTCLQNITL GKANKAFDYL 900
901 DAEADITSLT MFLYDNKEFV KLEQSLVLYF GDQLEKTFLP NLWKLHLGKD IDDSICLSEY 960
961 MPKNVINRVH NMWQKVMSQL EEDPFFKGMF ESTLGIPSSL PVIPSTMPNN ILKTPSKHST1020
1021 GLKLCDSPRS SSMTPRGKNI RQKFDRIAAI SKLKQMKELL ESLKLDTLDN HELSKISSLS1080
1081 SLTLTILSNI TSIHNAESSL ITNFSLTDLP RHMPLLFDKV LNNIDNKNYR EFRVSSLIAP1140
1141 NNISTITESI RVSAAQKDLM ESNLNINVIT IDFCPITGNL LLSKLEPRRK RRTHLRLPLI1200
1201 RSNSRDLDEV HLSFPEATKK LLSIINESNQ TTSVEVTNKI KTREERKSWW TTRYDLDKRM1260
1261 QQLLNNIENS WFNGVQGFFS PEVVDNSLFE KFKDKFYEIL HQNLPSRKLY GNPAMFIKVE1320
1321 DWVIELFLKL NPQEIDFLSK MEDLIYFVLD ILLFHGEENA YDEIDFSMLH VQLEEQIKKY1380
1381 RATMTTNSIF HTFLVVSSSC HLFPWECLSF LKDLSITRVP SYVCLNKLLS RFHYQLPLQV1440
1441 TIEDNISMIL NPNGDLSRTE SKFKGMFQKI IDAKPSSQLV MNEKPEEETL LKMLQNSNLF1500
1501 VYIGHGGGEQ YVRSKEIKKC TKIAPSFLLG CSSAAMKYYG KLEPTGTIYT YLLGGCPMVL1560
1561 GNLWDVTDKD IDKFSEELFE KMGFRCNTDD LNGNSLSVSY AVSKSRGVCH LRYLNGAAPV1620
1621 IYGLPIKFVS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
ESP1 PDS1 SRC1 |
| View Details | Krogan NJ, et al. (2006) |
|
CCL1 ESP1 KIN28 RAD3 SSL1 TFB1 TFB3 TFB4 |
| View Details | Gavin AC, et al. (2002) |
|
AMN1 ESP1 GTR2 RVB2 SPT16 YCH1 YCR015C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
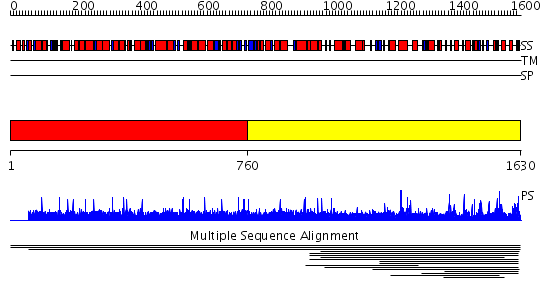
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..759] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [760..1630] | 1000.0 | Peptidase family C50 No confident structure predictions are available. | |
| 3 | View Details | [319..550] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [551..759] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [760..982] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [983..1186] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1187..1368] | 1.04098 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1369..1433] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1434..1521] | 1.047993 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1522..1630] | 1.047993 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)