













| General Information: |
|
| Name(s) found: |
XRS2 /
YDR369C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 854 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA Mre11 complex [IPI |
| Biological Process: |
double-strand break repair via break-induced replication
[TAS telomere maintenance [IMP double-strand break repair via nonhomologous end joining [IMP sporulation resulting in formation of a cellular spore [IMP meiotic DNA double-strand break formation [IMP |
| Molecular Function: |
DNA binding
[IDA protein binding [IPI telomeric DNA binding [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MWVVRYQNTL EDGSISFISC CLQAFKTYSI GRSSKNPLII KNDKSISRQH ITFKWEINNS 60
61 SDLKHSSLCL VNKGKLTSLN KKFMKVGETF TINASDVLKS TIIELGTTPI RIEFEWINEV 120
121 WNIPPHLTQF RTMLSEYGIS TEISINDIPA NLMISDYPKS EDNSIRELYA LVSTIPMKKS 180
181 RFLMELCNTL LPTSKTNLKF DEMWNDMISN PEYNVFDFDP NILLSKFMRL NNIRVLTTIK 240
241 SEPRLSSLLR TFNINLFAFD NIDSLYKYVD SLEASTEYLI LTTTDKKENG KILCTIKTML 300
301 TSIIDGTLSA VINMKGASSR TLDNGKFDQI SEGMSTILKT SRAPEVEASP VVSKKRKLNR 360
361 RRVLPLDSLD FFAGGLSTKT LSENRSLTDA KRLNCGAESK TVISSPNIAE ADEKHAPFLQ 420
421 NALKPTEDIG KKSGHSSPGA IIVSSPNLGT VNTSEDSLDK SLQSHKLPQP SLPEVAGIGS 480
481 QTISSNSADY ETAAVNSMDD AEVTKNFRVN HHQNIEQPSK NIRKLSNYSR EISSPLQENC 540
541 KSPVKELSIK EKSGTPHAFV EAIQETKNRE VKRVKSTIVE LKDEELSEEA INQLKNLAIV 600
601 EPSNNLLRKS FDSEGNKTSR TTEKWENSLM EPEWHKRKNF KTFVKVRPKS KAHKEEGKNN 660
661 TQSSDFIRNA AFLITRNYVP LKKYSKKDTT TKWGTEENED MFALTEMERF GSNTFMSDNI 720
721 NSNTIQKRSQ ALNRFTNEDS SNEGEEDSFS FSRCSGTAAS VQPLKNKIFI TDEDDLGDID 780
781 DKSDRLNHRE NNRNLFVVKE MNLRPNLSEE CSKQSRHSRS ATSRSRGSFG ASNNGDGDDD 840
841 DDDGPKFTFK RRKG |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
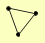
MRE11 XRS2 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
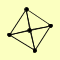
MRE11 RAD50 XRS2 |
| View Details | Krogan NJ, et al. (2006) |
|
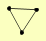
DMC1 MRE11 RAD50 XRS2 YBL044W |
| View Details | Ho Y, et al. (2002) |
|
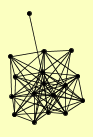
AHP1 CDC16 DUN1 ERG20 GPH1 MAM33 MKT1 MRE11 PST2 RAD50 REX2 RPT3 SEC27 TFP1 VMA8 XRS2 YBR063C |
| View Details | Qiu et al. (2008) |
|
MRE11 RAD50 XRS2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
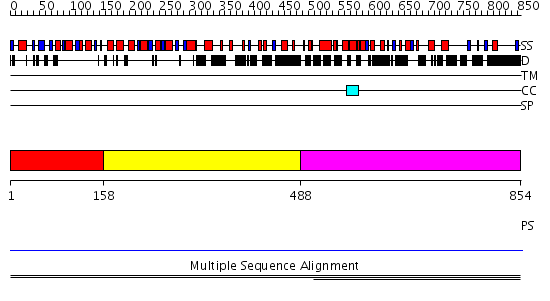
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..157] | 9.468521 | FHA domain No confident structure predictions are available. | |
| 2 | View Details | [158..487] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [488..854] | 1.002821 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [498..716] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [717..854] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)