













| General Information: |
|
| Name(s) found: |
CDC24 /
YAL041W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 854 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA incipient cellular bud site [IDA cellular bud neck [IDA mating projection tip [IDA |
| Biological Process: |
budding cell isotropic bud growth
[IPI small GTPase mediated signal transduction [TAS regulation of exit from mitosis [IMP pseudohyphal growth [IPI chitin localization [IMP invasive growth in response to glucose limitation [IPI cell morphogenesis involved in conjugation with cellular fusion [IMP budding cell apical bud growth [IPI pheromone-dependent signal transduction involved in conjugation with cellular fusion [IGI |
| Molecular Function: |
Rho guanyl-nucleotide exchange factor activity
[IDA signal transducer activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAIQTRFASG TSLSDLKPKP SATSISIPMQ NVMNKPVTEQ DSLFHICANI RKRLEVLPQL 60
61 KPFLQLAYQS SEVLSERQSL LLSQKQHQEL LKSNGANRDS SDLAPTLRSS SISTATSLMS 120
121 MEGISYTNSN PSATPNMEDT LLTFSMGILP ITMDCDPVTQ LSQLFQQGAP LCILFNSVKP 180
181 QFKLPVIASD DLKVCKKSIY DFILGCKKHF AFNDEELFTI SDVFANSTSQ LVKVLEVVET 240
241 LMNSSPTIFP SKSKTQQIMN AENQHRHQPQ QSSKKHNEYV KIIKEFVATE RKYVHDLEIL 300
301 DKYRQQLLDS NLITSEELYM LFPNLGDAID FQRRFLISLE INALVEPSKQ RIGALFMHSK 360
361 HFFKLYEPWS IGQNAAIEFL SSTLHKMRVD ESQRFIINNK LELQSFLYKP VQRLCRYPLL 420
421 VKELLAESSD DNNTKELEAA LDISKNIARS INENQRRTEN HQVVKKLYGR VVNWKGYRIS 480
481 KFGELLYFDK VFISTTNSSS EPEREFEVYL FEKIIILFSE VVTKKSASSL ILKKKSSTSA 540
541 SISASNITDN NGSPHHSYHK RHSNSSSSNN IHLSSSSAAA IIHSSTNSSD NNSNNSSSSS 600
601 LFKLSANEPK LDLRGRIMIM NLNQIIPQNN RSLNITWESI KEQGNFLLKF KNEETRDNWS 660
661 SCLQQLIHDL KNEQFKARHH SSTSTTSSTA KSSSMMSPTT TMNTPNHHNS RQTHDSMASF 720
721 SSSHMKRVSD VLPKRRTTSS SFESEIKSIS ENFKNSIPES SILFRISYNN NSNNTSSSEI 780
781 FTLLVEKVWN FDDLIMAINS KISNTHNNNI SPITKIKYQD EDGDFVVLGS DEDWNVAKEM 840
841 LAENNEKFLN IRLY |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
BOI2 CDC24 |
| View Details | Gavin AC, et al. (2006) |

|
BOI2 CDC24 |
| View Details | Gavin AC, et al. (2002) |
|
BEM1 BOI2 CDC24 HSP42 RSC2 SIS1 |
| View Details | Qiu et al. (2008) |
|
BEM1 BNI1 BUD5 CDC24 CDC42 FAR1 FUS3 GIC1 GIC2 LRG1 MOB2 RHO1 ROM2 RSR1 SHO1 SKT5 STE4 STE5 TPM2 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #24 Asynchronous Prep3-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
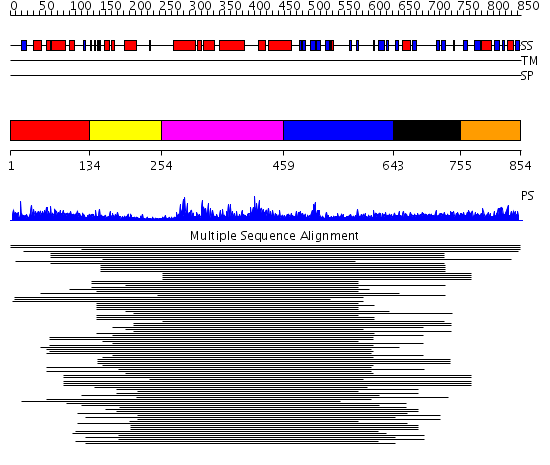
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 1.108997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [134..253] | 11.69897 | Calponin | |
| 3 | View Details | [254..458] | 202.457575 | GEF of TIAM1 (T-Lymphoma invasion and metastasis indusing protein 1) | |
| 4 | View Details | [459..642] | 202.457575 | GEF of TIAM1 (T-Lymphoma invasion and metastasis indusing protein 1) | |
| 5 | View Details | [643..754] | 1.061996 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [755..854] | N/A | Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||
| 5 |
|
|||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)