













| General Information: |
|
| Name(s) found: |
GIC1 /
YHR061C
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 314 amino acids |
Gene Ontology: |
|
| Cellular Component: |
incipient cellular bud site
[IDA cellular bud tip [IDA cellular bud neck [IDA mating projection tip [IDA actin cap [IDA |
| Biological Process: |
axial cellular bud site selection
[IMP regulation of exit from mitosis [IMP Rho protein signal transduction [IPI |
| Molecular Function: |
small GTPase regulator activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTEGKRLQQM ELPQMKSIWI DEDQEMEKLY GFQVRQRFMN GPSTDSDEDA DEDLGIVLVD 60
61 SKKLALPNKN NIKLPPLPNY MTINPNINSN HKSLTNKKKN FLGMFKKKDL LSRRHGSAKT 120
121 AKQSSISTPF DFHHISHANG KREDNPLESH EEKHDVESLV KFTSLAPQPR PDSNVSSKYS 180
181 NVVMNDSSRI VSSSTIATTM DSHHDGNETN NTPNGNKQLD SPTDLEMTLE DLRNYTFPSV 240
241 LGDSVSEKTN PSSPSVSSFS GKFKPRELSA LHTPELGNCF NVDQSLNSPG NRISVDDVLK 300
301 FYYQCSETST PRNT |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
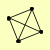
|
FIG2 GIC1 MRP13 YIL067C |
| View Details | Qiu et al. (2008) |
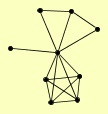
|
BFA1 BUD5 BUD6 CDC24 GIC1 MOB2 PEA2 PFY1 SLG1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
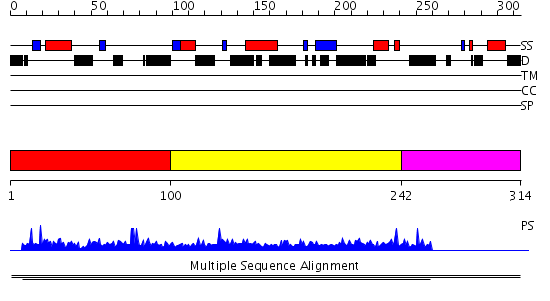
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [100..241] | 17.376751 | P21-Rho-binding domain No confident structure predictions are available. | |
| 3 | View Details | [242..314] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)