













| General Information: |
|
| Name(s) found: |
GAS1 /
YMR307W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 559 amino acids |
Gene Ontology: |
|
| Cellular Component: |
fungal-type cell wall
[IDA mitochondrion [IDA plasma membrane [IDA |
| Biological Process: |
filamentous growth
[IMP cellular cell wall organization [IMP |
| Molecular Function: |
1,3-beta-glucanosyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLFKSLSKLA TAAAFFAGVA TADDVPAIEV VGNKFFYSNN GSQFYIRGVA YQADTANETS 60
61 GSTVNDPLAN YESCSRDIPY LKKLNTNVIR VYAINTTLDH SECMKALNDA DIYVIADLAA 120
121 PATSINRDDP TWTVDLFNSY KTVVDTFANY TNVLGFFAGN EVTNNYTNTD ASAFVKAAIR 180
181 DVRQYISDKN YRKIPVGYSS NDDEDTRVKM TDYFACGDDD VKADFYGINM YEWCGKSDFK 240
241 TSGYADRTAE FKNLSIPVFF SEYGCNEVTP RLFTEVEALY GSNMTDVWSG GIVYMYFEET 300
301 NKYGLVSIDG NDVKTLDDFN NYSSEINKIS PTSANTKSYS ATTSDVACPA TGKYWSAATE 360
361 LPPTPNGGLC SCMNAANSCV VSDDVDSDDY ETLFNWICNE VDCSGISANG TAGKYGAYSF 420
421 CTPKEQLSFV MNLYYEKSGG SKSDCSFSGS ATLQTATTQA SCSSALKEIG SMGTNSASGS 480
481 VDLGSGTESS TASSNASGSS SKSNSGSSGS SSSSSSSSAS SSSSSKKNAA TNVKANLAQV 540
541 VFTSIISLSI AAGVGFALV |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Ho Y, et al. (2002) |
|
ABP1 ACO1 ADH6 AIM41 ATP4 BUD14 CAR2 COR1 CRZ1 CYS4 DCP1 DCP2 DNM1 EDC3 EDE1 EGD2 ENP1 GAS1 GCN3 GPH1 HHF1, HHF2 HRR25 HSP104 HXT7 HYP2 ILV5 IPP1 LOC1 LSP1 LTV1 MDH1 MDS3 MGE1 NSA1 NUG1 OYE2 PEX19 PIL1 PIN4 PTC4 PUF3 RIO2 RNR2 RPC19 RPM2 SAH1 SAP185 SAP190 SAS10 SEC2 SEC23 SES1 SFB3 SGM1 SIR1 SIR3 SIT4 TEF4 TMA19 TMA29 TSR1 VMA4 YBR225W YER138C |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | his-HA tag on RPA135 | Schneider, DA, et al. (2006) | |
| View Run | Dam1-765 complex purified from yeast with Dad1-TAP | Shimogawa MM, et al. (2006) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | phosphorylation data - cascade search | Green EM, et al (2005) | |
| View Run | #04 Alpha Factor Prep2 | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
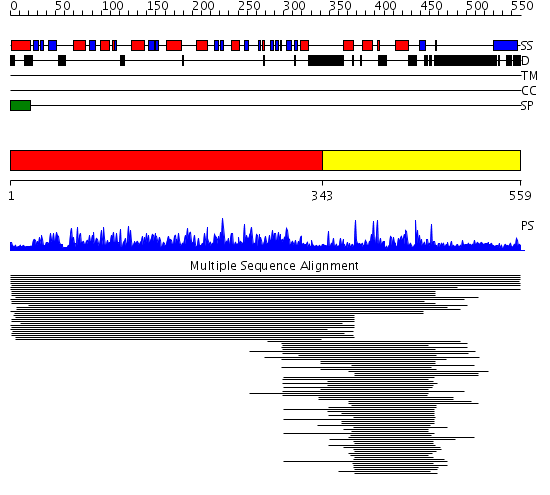
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..342] | 236.327902 | Glycolipid anchored surface protein (GAS1) No confident structure predictions are available. | |
| 2 | View Details | [343..559] | 44.092985 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|