













| General Information: |
|
| Name(s) found: |
MPK4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 30 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 376 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytoplasm [IDA] |
| Biological Process: |
hypotonic salinity response
[IDA]
response to salt stress [IDA] signal transduction [IC] systemic acquired resistance, salicylic acid mediated signaling pathway [IMP][TAS] response to fungus [IMP] jasmonic acid and ethylene-dependent systemic resistance [IMP] cortical microtubule organization [IMP] jasmonic acid and ethylene-dependent systemic resistance, jasmonic acid mediated signaling pathway [TAS] response to stress [TAS] hyperosmotic response [IMP] response to biotic stimulus [TAS] response to cold [IDA] response to abscisic acid stimulus [IEP] phosphorylation [IDA] |
| Molecular Function: |
MAP kinase activity
[IDA][ISS]
kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSAESCFGSS GDQSSSKGVA THGGSYVQYN VYGNLFEVSR KYVPPLRPIG RGAYGIVCAA 60
61 TNSETGEEVA IKKIGNAFDN IIDAKRTLRE IKLLKHMDHE NVIAVKDIIK PPQRENFNDV 120
121 YIVYELMDTD LHQIIRSNQP LTDDHCRFFL YQLLRGLKYV HSANVLHRDL KPSNLLLNAN 180
181 CDLKLGDFGL ARTKSETDFM TEYVVTRWYR APELLLNCSE YTAAIDIWSV GCILGETMTR 240
241 EPLFPGKDYV HQLRLITELI GSPDDSSLGF LRSDNARRYV RQLPQYPRQN FAARFPNMSA 300
301 GAVDLLEKML VFDPSRRITV DEALCHPYLA PLHDINEEPV CVRPFNFDFE QPTLTEENIK 360
361 ELIYRETVKF NPQDSV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
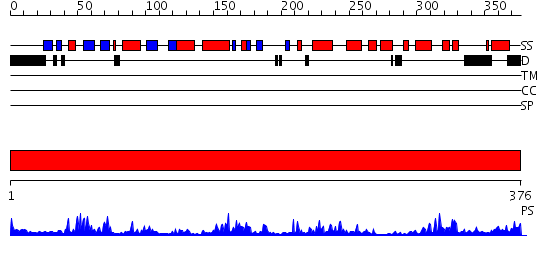
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..376] | 89.69897 | P38 MITOGEN-ACTIVATED KINASE IN COMPLEX WITH 4-AZAINDOLE INHIBITOR |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)