













| General Information: |
|
| Name(s) found: |
MPK3_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 370 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
camalexin biosynthetic process
[IMP]
signal transduction [IC] response to osmotic stress [IDA] response to wounding [IEP] priming of cellular response to stress [IMP] activation of MAPK activity involved in osmosensory signaling pathway [IDA] response to stress [TAS] response to oxidative stress [IEP][TAS] response to cold [IEP] abscisic acid mediated signaling pathway [TAS] response to bacterium [IEP] response to chitin [IEP] ovule development [IGI] |
| Molecular Function: |
protein binding
[IPI]
MAP kinase activity [ISS] protein kinase activity [TAS] kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNTGGGQYTD FPAVETHGGQ FISYDIFGSL FEITSKYRPP IIPIGRGAYG IVCSVLDTET 60
61 NELVAMKKIA NAFDNHMDAK RTLREIKLLR HLDHENIIAI RDVVPPPLRR QFSDVYISTE 120
121 LMDTDLHQII RSNQSLSEEH CQYFLYQLLR GLKYIHSANI IHRDLKPSNL LLNANCDLKI 180
181 CDFGLARPTS ENDFMTEYVV TRWYRAPELL LNSSDYTAAI DVWSVGCIFM ELMNRKPLFP 240
241 GKDHVHQMRL LTELLGTPTE SDLGFTHNED AKRYIRQLPN FPRQPLAKLF SHVNPMAIDL 300
301 VDRMLTFDPN RRITVEQALN HQYLAKLHDP NDEPICQKPF SFEFEQQPLD EEQIKEMIYQ 360
361 EAIALNPTYG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
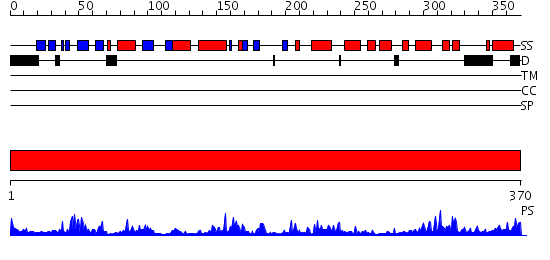
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..370] | 93.39794 | No description for 2gtmA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)