













| General Information: |
|
| Name(s) found: |
atp-2 /
CE29950
[WormBase]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Caenorhabditis elegans |
| Length: | 538 amino acids |
Gene Ontology: |
|
| Cellular Component: |
proton-transporting two-sector ATPase complex
[IEA bacterial-type flagellum [IEA cytoplasm [IEA mitochondrion [IDA cilium [IDA integral to membrane [IEA |
| Biological Process: |
embryonic development ending in birth or egg hatching
[IMP positive regulation of growth rate [IMP protein transport [IEA flagellum assembly [IEA transcription termination [IEA biosynthetic process [IEA ATP biosynthetic process [IEA reproduction [IMP growth [IMP mating behavior [IMP nematode larval development [IMP ciliary or flagellar motility [IEA protein secretion by the type III secretion system [IEA energy coupled proton transport, against electrochemical gradient [IEA ATP synthesis coupled proton transport [IEA |
| Molecular Function: |
nucleotide binding
[IEA ATPase activity [IEA ATP binding [IEA protein domain specific binding [IPI transcription termination factor activity [IEA nucleoside-triphosphatase activity [IEA proton-transporting ATPase activity, rotational mechanism [IEA hydrogen-exporting ATPase activity, phosphorylative mechanism [IEA hydrogen ion transporting ATP synthase activity, rotational mechanism [IEA hydrolase activity, acting on acid anhydrides, catalyzing transmembrane movement of substances [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASRSLASIS RSASRLLQSN VQKCALPAAS IRLSSNNVES KKGIHTGVAT QQAAAATKVS 60
61 AKATAANASG RIVAVIGAVV DVQFDENLPP ILNGLEVVGR SPRLILEVSQ HLGDNVVRCI 120
121 AMDGTEGLVR GQPVADTGDP IKIPVGPETL GRIMNVIGEP IDERGPIASK NFAAIHAEAP 180
181 EFVEMSVEQE ILVTGIKVVD LLAPYAKGGK IGLFGGAGVG KTVLIMELIN NVAKAHGGYS 240
241 VFAGVGERTR EGNDLYHEMI EGGVIDLKGK NSKVSLVYGQ MNEPPGARAR VCLTGLTVAE 300
301 YFRDQEGQDV LLFIDNIFRF TQAGSEVSAL LGRIPSAVGY QPTLATDMGS MQERITTTKK 360
361 GSITSVQAIY VPADDLTDPA PATTFAHLDA TTVLSRGIAE LAIYPAVDPL DSTSRIMDPN 420
421 VVGQNHYDIA RGVQKILQDY KSLQDIIAIL GMDELSEEDK LTVSRARKIQ RFLSQPFQVA 480
481 EVFTGHQGKF VSLEETIRGF TMILKGELDH LPEVAFYMQG GIDDVFKKAE ELAKQHGN |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | hcp1 IP from december2004 | Cheeseman, I.M., et al. (2005) | |
| View Run | hcp2 ip from december | Cheeseman, I.M., et al. (2005) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
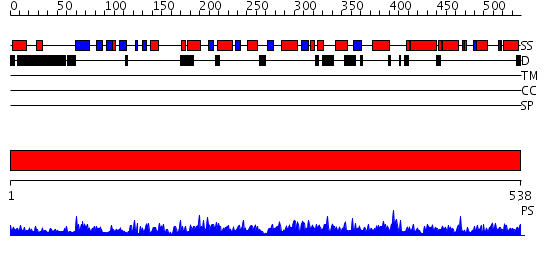
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..538] | 1000.0 | F1 ATP synthase beta subunit, domain 3; F1 ATP synthase beta subunit, domain 1; Central domain of beta subunit of F1 ATP synthase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.57 |
Source: Reynolds et al. (2008)