













| General Information: |
|
| Name(s) found: |
SAP155 /
YFR040W
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1002 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IPI |
| Biological Process: |
G1/S transition of mitotic cell cycle
[IGI |
| Molecular Function: |
protein serine/threonine phosphatase activity
[IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSFWPFGQNL NHSNINKILD EYFHVLHELE RINPSVGKAI PAIFNNVQER GTSDSLDSIP 60
61 EEYSHGDEVK TARGDQKSRF EKDDQQERYE KEEEERSMNS SESSTTSFSS GSTSKTDLDE 120
121 EDISNATAPM MVTTKNLDNS FIERMLVETE LLNELSRQNK TLLDFICFGF FFDKKTNKKV 180
181 NNMEYLVDQL MECISKIKTA TTVDLNNLID YQEQQQLDDS SQEDVYVESD TEQEEEKEDD 240
241 NNSNNKKRRK RGSSSFGNDD INNNDDDDDA NEDDESAYLT KATIISEIFS LDIWLISESL 300
301 VKNQSYLNKI WSIINQPNFN SENSPLVPIF LKINQNLLLT RQDQYLNFIR TERSFVDDML 360
361 KHVDISLLMD FFLKIISTDK IESPTGIIEL VYDQNLISKC LSFLNNKESP ADIQACVGDF 420
421 LKALIAISAN APLDDISIGP NSLTRQLASP ESIAKLVDIM INQRGAALNT TVSIVIELIR 480
481 KNNSDYDQVN LLTTTIKTHP PSNRDPIYLG YLLRKFSNHL SDFFQIILDI ENDANIPLHE 540
541 NQLHEKFKPL GFERFKVVEL IAELLHCSNM GLMNSKRAER IARRRDKVRS QLSHHLQDAL 600
601 NDLSIEEKEQ LKTKHSPTRD TDHDLKNNNG KIDNDNNDND DESDYGDEID ESFEIPYINM 660
661 KQTIKLRTDP TVGDLFKIKL YDTRIVSKIM ELFLTHPWNN FWHNVIFDII QQIFNGRMDF 720
721 SYNSFLVLSL FNLKSSYQFM TDIVISDEKG TDVSRFSPVI RDPNFDFKIT TDFILRGYQD 780
781 SYKFYELRKM NLGYMGHIVL IAEEVVKFSK LYKVELISPD IQVILQTEEW QYYSEEVLNE 840
841 TRMMYSKILG GGSYIDDGNG NIIPQLPDNT TVLTPNGDAS NNNEILDSDT GSSNGTSGGG 900
901 QLINVESLEE QLSLSTESDL HNKLREMLIN RAQEDVDNKN TENGVFILGP PEDKNSNSNI 960
961 NNTNHNSNNS NNNDNNDNND NDNDNTRNYN EDADNDNDYD HE |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ATG18 SAP155 SAP190 SIT4 |
| View Details | Krogan NJ, et al. (2006) |
|
HRT3 PPZ2 SAP155 SDS24 SIT4 |
| View Details | Gavin AC, et al. (2002) |
|
ACC1 AVT4 BEM2 CCT2 CDC25 FAB1 HRK1 MDS3 MIA40 MRPL3 SAP155 SAP185 SAP190 SIT4 SWT21 |
| View Details | Ho Y, et al. (2002) |

|
ACC1 ADO1 ARP2 ATG18 ATG2 ATG20 ATP3 BGL2 CCT6 CDC55 FAA1 FAA4 GLT1 GSF2 HXT5 MAE1 NDE1 NPA3 PFK2 RMT2 RNR2 RPT4 SAP155 SAP185 SAP190 SIT4 TAP42 THI21 TIP41 UTP13 YMR196W |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #10 Mitotic Prep1-TiO2 Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
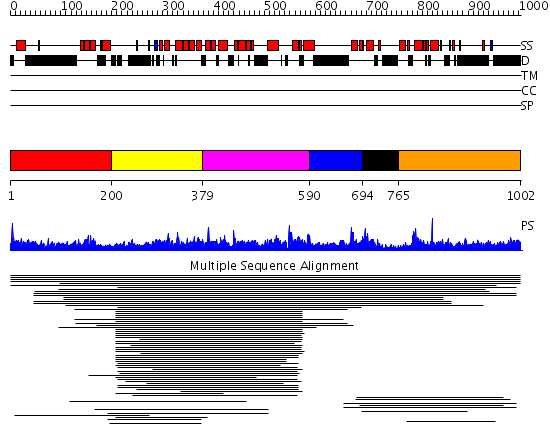
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [200..378] | 2.03997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [379..589] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [590..693] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [694..764] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [765..1002] | 1.015972 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)