













| General Information: |
|
| Name(s) found: |
LAP3 /
YNL239W
[SGD]
|
| Description(s) found:
Found 22 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 454 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA mitochondrion [IDA |
| Biological Process: |
response to antibiotic
[IDA |
| Molecular Function: |
nucleic acid binding
[TAS transcription regulator activity [IDA cysteine-type peptidase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSSIDISKI NSWNKEFQSD LTHQLATTVL KNYNADDALL NKTRLQKQDN RVFNTVVSTD 60
61 STPVTNQKSS GRCWLFAATN QLRLNVLSEL NLKEFELSQA YLFFYDKLEK ANYFLDQIVS 120
121 SADQDIDSRL VQYLLAAPTE DGGQYSMFLN LVKKYGLIPK DLYGDLPYST TASRKWNSLL 180
181 TTKLREFAET LRTALKERSA DDSIIVTLRE QMQREIFRLM SLFMDIPPVQ PNEQFTWEYV 240
241 DKDKKIHTIK STPLEFASKY AKLDPSTPVS LINDPRHPYG KLIKIDRLGN VLGGDAVIYL 300
301 NVDNETLSKL VVKRLQNNKA VFFGSHTPKF MDKKTGVMDI ELWNYPAIGY NLPQQKASRI 360
361 RYHESLMTHA MLITGCHVDE TSKLPLRYRV ENSWGKDSGK DGLYVMTQKY FEEYCFQIVV 420
421 DINELPKELA SKFTSGKEEP IVLPIWDPMG ALAK |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
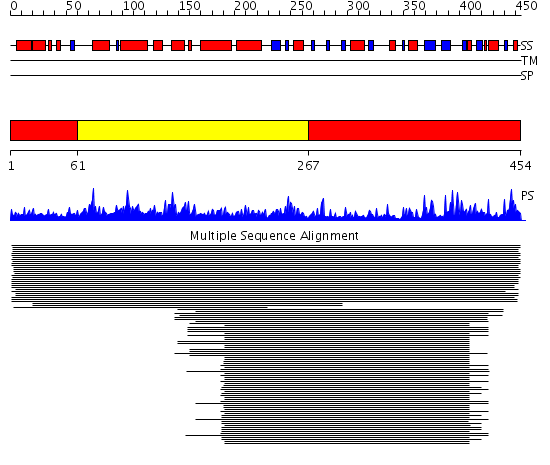
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..60] [267..454] |
10154.0 | Bleomycin hydrolase | |
| 2 | View Details | [61..266] | 10154.0 | Bleomycin hydrolase |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)