













| General Information: |
|
| Name(s) found: |
HSP82 /
YPL240C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 709 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
response to osmotic stress
[IMP protein refolding [IMP proteasome assembly [IDA 'de novo' protein folding [IDA response to stress [IMP |
| Molecular Function: |
unfolded protein binding
[IDA ATPase activity, coupled [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASETFEFQA EITQLMSLII NTVYSNKEIF LRELISNASD ALDKIRYKSL SDPKQLETEP 60
61 DLFIRITPKP EQKVLEIRDS GIGMTKAELI NNLGTIAKSG TKAFMEALSA GADVSMIGQF 120
121 GVGFYSLFLV ADRVQVISKS NDDEQYIWES NAGGSFTVTL DEVNERIGRG TILRLFLKDD 180
181 QLEYLEEKRI KEVIKRHSEF VAYPIQLVVT KEVEKEVPIP EEEKKDEEKK DEEKKDEDDK 240
241 KPKLEEVDEE EEKKPKTKKV KEEVQEIEEL NKTKPLWTRN PSDITQEEYN AFYKSISNDW 300
301 EDPLYVKHFS VEGQLEFRAI LFIPKRAPFD LFESKKKKNN IKLYVRRVFI TDEAEDLIPE 360
361 WLSFVKGVVD SEDLPLNLSR EMLQQNKIMK VIRKNIVKKL IEAFNEIAED SEQFEKFYSA 420
421 FSKNIKLGVH EDTQNRAALA KLLRYNSTKS VDELTSLTDY VTRMPEHQKN IYYITGESLK 480
481 AVEKSPFLDA LKAKNFEVLF LTDPIDEYAF TQLKEFEGKT LVDITKDFEL EETDEEKAER 540
541 EKEIKEYEPL TKALKEILGD QVEKVVVSYK LLDAPAAIRT GQFGWSANME RIMKAQALRD 600
601 SSMSSYMSSK KTFEISPKSP IIKELKKRVD EGGAQDKTVK DLTKLLYETA LLTSGFSLDE 660
661 PTSFASRINR LISLGLNIDE DEETETAPEA STAAPVEEVP ADTEMEEVD |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CPR6 FBA1 HSC82 HSP104 HSP82 STI1 TIF1, TIF2 YEF3 |
| View Details | Krogan NJ, et al. (2006) |

|
AHA1 BUD3 CNS1 CPR6 DIG2 DNM1 DSF1, YNR073C FRT2 GPA2 HOM6 HSC82 HSP104 HSP82 MBB1 MRPL50 PAM1 PPT1 QNS1 RMD8 RTS1 SMT3 STI1 STP4 TDH1 UTP20 YBL104C |
| View Details | Gavin AC, et al. (2002) |
|
HSC82 HSP82 PPT1 |
The following runs contain data for this protein:
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
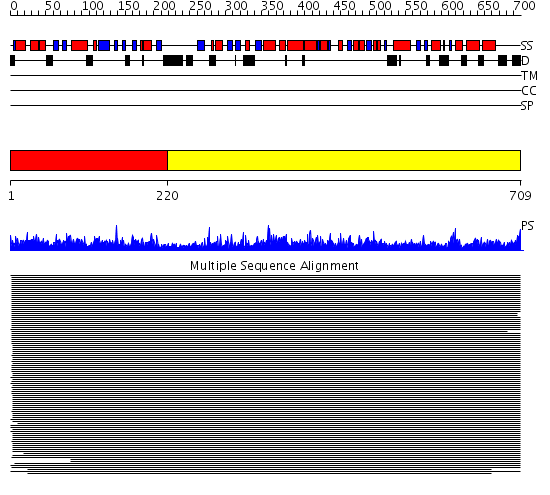
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..219] | 1236.39794 | HSP90 | |
| 2 | View Details | [220..709] | 1000.0 | Hsp90 protein No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)