













| General Information: |
|
| Name(s) found: |
ATG26 /
YLR189C
[SGD]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1198 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA |
| Biological Process: |
sterol metabolic process
[IMP |
| Molecular Function: |
sterol 3-beta-glucosyltransferase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPITQIISAS DSEAGPKPSI SLVPDKPSEP ETSPRHHRLS RSLSKFKRWR GRSNSSLSMG 60
61 SSEQQELQDS PNEARSDDDE NGYNNDNADD LAKSKYMMKS IAGLLTTASV YAGMNNAQEM 120
121 NVLSQVDSEE SDSSDSFQEN IGRNEVKSKK ENLKTKSHPE VPRLDKRKPT LFDFSITREK 180
181 LSKDNVAKLR QRFCLDEQEP FLNDFPAWLL KDVLVQGHIF ITTKHFLFFA YLPKNPRSVK 240
241 MSGNLNIRTK LIRSTRYWCV LKNHLFSMYT SSTELYFPVL TIDLREVQKI ETQKHTLNGS 300
301 ATKTFKLYTD ESTFKFNADS EFSAKSWVNA LKKEQFAAQN SENNSISLKI PLPNIIEIDD 360
361 QPIVNKALTL RLRALESSQT YAIDDFMFVF MDGSGSQVKE SLGEQLAILQ KSGVNTLYYD 420
421 IPAKKSKSSF GKETPATVEQ KNNGEDSKYL NVPTSAVPSS ENGKKSRFRF RERSNSWFRR 480
481 AKPLEDSQVE DVEEIYKDAA NDIDSSVHST IHIHEQEDSQ EQTVAWKPSH LKNFAEMWAA 540
541 KPIHYRNKFI PFQKDDTYLI KETEEVSANE RFRYHFKFNK EKSLISTYYT YLNRNVPVYG 600
601 KIYVSNDTVC FRSLLPGSNT YMVLPLVDVE TCYKEKGFRF GYFVLVIVIH GHEELFFEFS 660
661 TEVARDDIER ILLKLLDNIY ASSAEGSNIS SASLGDVQHN PDSAKLKLFE DKINAEGFEV 720
721 PLMIDENPHY KTSIKPNKSY KFGLLTIGSR GDVQPYIALG KGLIKEGHQV VIITHSEFRD 780
781 FVESHGIQFE EIAGNPVELM SLMVENESMN VKMLREASSK FRGWIDALLQ TSWEVCNRRK 840
841 FDILIESPSA MVGIHITEAL QIPYFRAFTM PWTRTRAYPH AFIVPDQKRG GNYNYLTHVL 900
901 FENVFWKGIS GQVNKWRVET LGLGKTNLFL LQQNNVPFLY NVSPTIFPPS IDFSEWVRVT 960
961 GYWFLDDKST FKPPAELQEF ISEARSKGKK LVYIGFGSIV VSNAKEMTEA LVEAVMEADV1020
1021 YCILNKGWSE RLGDKAAKKT EVDLPRNILN IGNVPHDWLF PQVDAAVHHG GSGTTGASLR1080
1081 AGLPTVIKPF FGDQFFYAGR VEDIGVGIAL KKLNAQTLAD ALKVATTNKI MKDRAGLIKK1140
1141 KISKEDGIKT AISAIYNELE YARSVTLSRV KTPRKKEENV DATKLTPAET TDEGWTMI |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
ATG26 MRPL20 MRPL28 MRPL8 |
| View Details | Gavin AC, et al. (2006) |
|
ATG26 BUD3 MRPL10 MRPL16 MRPL23 MRPL4 MRPL51 |
| View Details | Gavin AC, et al. (2002) |

|
ATG26 BEM2 BUD3 COP1 CYR1 HRR25 IMG1 IMG2 IML1 MHR1 MRH4 MRM1 MRP20 MRP49 MRP7 MRPL1 MRPL10 MRPL13 MRPL16 MRPL17 MRPL19 MRPL20 MRPL23 MRPL24 MRPL25 MRPL27 MRPL28 MRPL3 MRPL35 MRPL36 MRPL39 MRPL4 MRPL44 MRPL51 MRPL6 MRPL7 MRPL8 NUG1 PRP43 RET2 ROM2 RTC6 SEC1 SEC26 SEC27 SIN4 SNF2 SWI3 SYP1 TAF5 UTP20 YDR115W YML6 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Cheeseman IM, et al. (2002) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #11 Mitotic Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
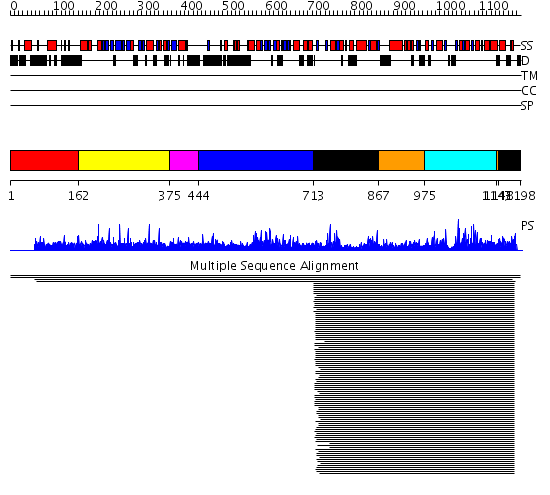
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [162..374] | 13.65 | Tapp1 | |
| 3 | View Details | [375..443] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [444..712] | 8.039965 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [713..866] [1148..1198] |
192.758605 | UDP-glucosyltransferase GtfB | |
| 6 | View Details | [867..974] [1143..1147] |
192.758605 | UDP-glucosyltransferase GtfB | |
| 7 | View Details | [975..1142] | 192.758605 | UDP-glucosyltransferase GtfB |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 |
|
||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)