













| General Information: |
|
| Name(s) found: |
DDI1 /
YER143W
[SGD]
|
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 428 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA |
| Biological Process: |
vesicle-mediated transport
[IGI ubiquitin-dependent protein catabolic process [IMP |
| Molecular Function: |
SNARE binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDLTISNELT GEIYGPIEVS EDMALTDLIA LLQADCGFDK TKHDLYYNMD ILDSNRTQSL 60
61 KELGLKTDDL LLIRGKISNS IQTDAATLSD EAFIEQFRQE LLNNQMLRSQ LILQIPGLND 120
121 LVNDPLLFRE RLGPLILQRR YGGYNTAMNP FGIPQDEYTR LMANPDDPDN KKRIAELLDQ 180
181 QAIDEQLRNA IEYTPEMFTQ VPMLYINIEI NNYPVKAFVD TGAQTTIMST RLAKKTGLSR 240
241 MIDKRFIGEA RGVGTGKIIG RIHQAQVKIE TQYIPCSFTV LDTDIDVLIG LDMLKRHLAC 300
301 VDLKENVLRI AEVETSFLSE AEIPKSFQEG LPAPTSVTTS SDKPLTPTKT SSTLPPQPGA 360
361 VPALAPRTGM GPTPTGRSTA GATTATGRTF PEQTIKQLMD LGFPRDAVVK ALKQTNGNAE 420
421 FAASLLFQ |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #34 Asynchronous SPB prep | Keck JM, et al. (2011) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
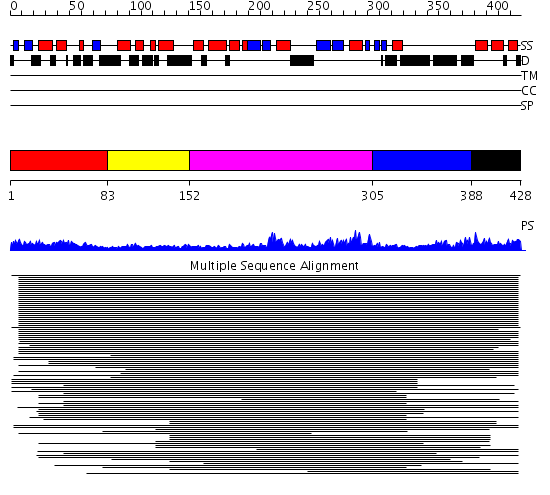
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 20.39794 | Ubiquitin | |
| 2 | View Details | [83..151] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [152..304] | 19.69897 | Human immunodeficiency virus type 1 protease | |
| 4 | View Details | [305..387] | 7.39794 | No description for 1rvnA_ was found. | |
| 5 | View Details | [388..428] | 4.522879 | DNA repair protein Hhr23a |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 |
|
||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)