













| General Information: |
|
| Name(s) found: |
UBX5 /
YDR330W
[SGD]
|
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 500 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
ubiquitin-dependent protein catabolic process
[IEP |
| Molecular Function: |
molecular_function
[ND]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEGKVDEFM AITGADDAAI ATQFIEMADG NLNTAISLFF ENGGAALLSS NNTPTPSNST 60
61 PMAPTSVDSD ADAQLAERLQ REAYQQQQPD QDYVRPPDEA RHEVLTETSG FPISYGGIGG 120
121 RFEPLHRVND MFDEGRPESI FNQRLDDTNT NTYINDNSSD SLDSEEENDD DEYEYVEEPV 180
181 IELDEDGNIK EYTKLVRKPK TISKEQKLAL LFRPPFSIMS KLDLDAAKQK ARAKQKWIMI 240
241 NIQDSGIFQC QALNRDLWSS RPVKTIIKEN FVFLQYQYES RNAQPYLQFY HLNNKDDLPH 300
301 IAILDPITGE RVKQWNRVVP IPEQFISEIN EFLASFSLDP KVPNPTVNEP LPKVDPTTLT 360
361 EEQQMELAIK ESLNNNSSKS NQEEVPSTGE EQKRVQEPDP FSTIEARVHP EPPNKPGITT 420
421 RIQIRTGDGS RLVRRFNALE DTVRTIYEVI KTEMDGFADS RFTLNDHQRE DLIDKLNMTI 480
481 ADAGLKNSSL LLEKLDPEIE |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
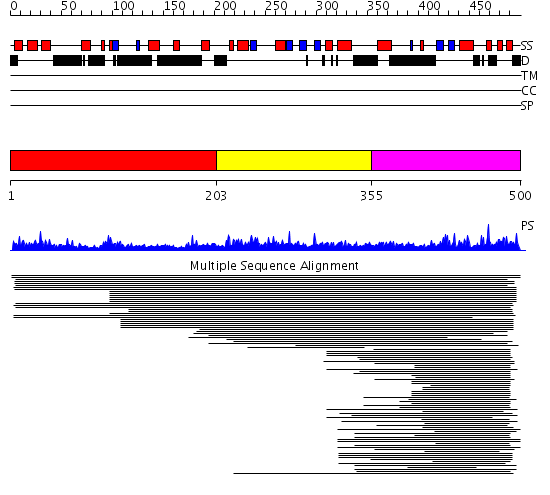
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..202] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [203..354] | 1.046973 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [355..500] | 19.221849 | p47 | |
| 4 | View Details | [383..500] | 20.39794 | Crystal structure of AAA ATPase p97/VCP ND1 in complex with p47 C |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)