













| General Information: |
|
| Name(s) found: |
FOB1 /
YDR110W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 566 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA |
| Biological Process: |
chromatin silencing at rDNA
[IDA extrachromosomal circular DNA accumulation involved in cell aging [IMP DNA recombination [IMP negative regulation of DNA replication [IDA replicative cell aging [IMP |
| Molecular Function: |
rDNA binding
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTKPRYNDVL FDDDDSVPSE SVTRKSQRRK ATSPGESRES SKDRLLILPS MGESYTEYVD 60
61 SYLNLELLER GERETPIFLE SLTRQLTQKI YELIKTKSLT ADTLQQISDK YDGVVAENKL 120
121 LFLQRQYYVD DEGNVRDGRN NDKIYCEPKH VYDMVMATHL MNKHLRGKTL HSFLFSHFAN 180
181 ISHAIIDWVQ QFCSKCNKKG KIKPLKEYKR PDMYDKLLPM ERIHIEVFEP FNGEAIEGKY 240
241 SYVLLCRDYR SSFMWLLPLK STKFKHLIPV VSSLFLTFAR VPIFVTSSTL DKDDLYDICE 300
301 EIASKYGLRI GLGLKSSARF HTGGILCIQY ALNSYKKECL ADWGKCLRYG PYRFNRRRNK 360
361 RTKRKPVQVL LSEVPGHNAK FETKRERVIE NTYSRNMFKM AGGKGLIYLE DVNTFALANE 420
421 ADNSCNNNGI LHNNNIGNDN FEEEVQKQFD LTEKNYIDEY DDLAHDSSEG EFEPNTLTPE 480
481 EKPPHNVDED RIESTGVAAP MQGTEEPEKG DQKESDGASQ VDQSVEITRP ETSYYQTLES 540
541 PSTKRQKLDQ QGNGDQTRDF GTSMEL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |
|
FOB1 NET1 NSR1 TOP1 UTP9 |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
| View Data |
|
Huh WK, et al. (2003) |
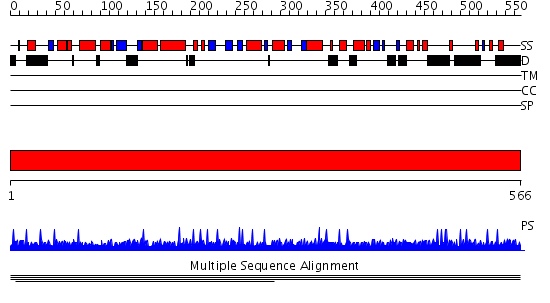
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..566] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [131..484] | 5.37 | N-terminal Zn binding domain of HIV integrase; Retroviral integrase, catalytic domain | |
| 3 | View Details | [485..566] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.89 |
Source: Reynolds et al. (2008)