













| General Information: |
|
| Name(s) found: |
sgk-1 /
CE39527
[WormBase]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Caenorhabditis elegans |
| Length: | 453 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
dauer larval development
[IMP cell communication [IEA embryonic development ending in birth or egg hatching [IMP determination of adult lifespan [IMP oviposition [IMP protein amino acid phosphorylation [IEA |
| Molecular Function: |
ATP binding
[IEA protein tyrosine kinase activity [IEA zinc ion binding [IEA protein binding [IEA phosphoinositide binding [IEA protein kinase activity [IEA protein serine/threonine kinase activity [IEA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVRKDEVTCN VIIGDDKKTV VYALRIGNGP IMQKTFEEYE RFFTTEKDMI PATIFTAPKK 60
61 KFLQADSKFY EKRRVWILVI SQHLVDNNLR SEDVRRFFHL ESPDDDENNV DLGPSERKTA 120
121 TANDFDYLTT IGKGSFGRVY QVRHKETKKI YAMKILSKEH IRKKNEVKHV MAERNVLINN 180
181 FKHPFLVSLH FSFQNKEKLY FVLDHLNGGE LFSHLQREKH FSESRSRFYA AEIACALGYL 240
241 HEKNIIYRDL KPENLLLDDK GYLVLTDFGL CKEDMQGSKT TSTFCGTPEY LAPEIILKKP 300
301 YDKTVDWWCL GSVLYEMIFG LPPFYSKDHN EMYDKIINQP LRLKHNISVP CSELITGLLQ 360
361 KDRSKRLGHR NDFRDIRDHP FFLPVDWDKL LNRELKAPFI PKVKNAMDTS NISKEFVEIQ 420
421 IDPSSLAPQQ LAVTHRDHDF ENFTFVDTNR VLV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
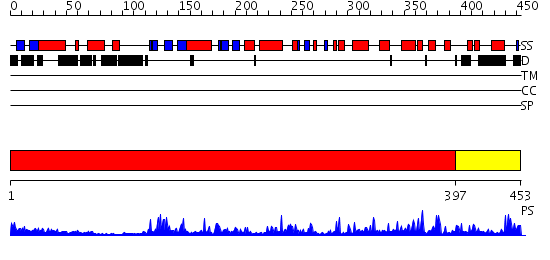
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..396] | 54.0 | CHICKEN SRC TYROSINE KINASE | |
| 2 | View Details | [397..453] | 93.39794 | crystal structure of an inactive Akt2 kinase domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)