













| General Information: |
|
| Name(s) found: |
SST2 /
YLR452C
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 698 amino acids |
Gene Ontology: |
|
| Cellular Component: |
plasma membrane
[IDA |
| Biological Process: |
signal transduction
[IMP adaptation of signaling pathway by response to pheromone involved in conjugation with cellular fusion [IMP |
| Molecular Function: |
GTPase activator activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVDKNRTLHE LSSKNFSRTP NGLIFTNDLK TVYSIFLICL DLKEKKHSSD TKSFLLTAFT 60
61 KHFHFTFTYQ EAIKAMGQLE LKVDMNTTCI NVSYNIKPSL ARHLLTLFMS SKLLHTPQDR 120
121 TRGEPKEKVL FQPTPKGVAV LQKYVRDIGL KTMPDILLSS FNSMKLFTFE RSSVTDSIIH 180
181 SDYLIHILFI KMMGAKPNVW SPTNADDPLP CLSSLLEYTN NDDTFTFEKS KPEQGWQAQI 240
241 GNIDINDLER VSPLAHRFFT NPDSESHTQY YVSNAGIRLF ENKTFGTSKK IVIKYTFTTK 300
301 AIWQWIMDCT DIMHVKEAVS LAALFLKTGL IVPVLLQPSR TDKKKFQISR SSFFTLSKRG 360
361 WDLVSWTGCK SNNIRAPNGS TIDLDFTLRG HMTVRDEKKT LDDSEGFSQD MLISSSNLNK 420
421 LDYVLTDPGM RYLFRRHLEK ELCVENLDVF IEIKRFLKKM TILKKLIDSK HCDKKSNTST 480
481 SKNNIVKTID SALMKQANEC LEMAYHIYSS YIMIGSPYQL NIHHNLRQNI SDIMLHPHSP 540
541 LSEHFPTNLY DPSPASAESA ASSISSTEAD TLGEPPEVSL KPSKNLSNEN CSFKKQGFKH 600
601 QLKEYKPAPL TLAETHSPNA SVENSHTIVR YGMDNTQNDT KSVESFPATL KVLRKLYPLF 660
661 EIVSNEMYRL MNNDSFQKFT QSDVYKDASA LIEIQEKC |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Hazbun TR, et al. (2003) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
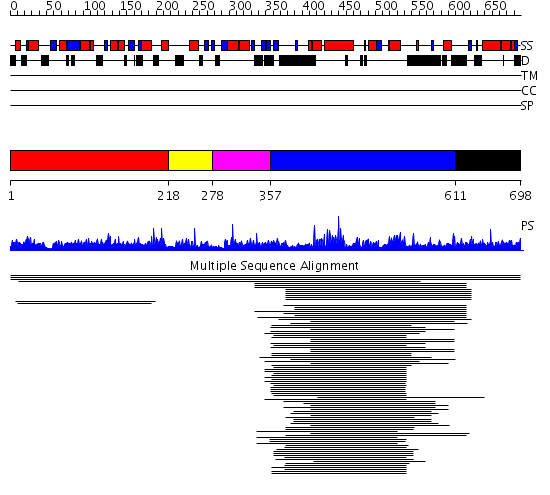
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..217] | 2.00494 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [218..277] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [278..356] | 15.920819 | Domain found in Dishevelled, Egl-10, and Pleckstrin No confident structure predictions are available. | |
| 4 | View Details | [357..610] | 48.69897 | Regulator of G-protein signalling 4, RGS4 | |
| 5 | View Details | [611..698] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)