













| General Information: |
|
| Name(s) found: |
PEP12 /
YOR036W
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 288 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endosome
[IMP Golgi apparatus [IMP |
| Biological Process: |
Golgi to vacuole transport
[IMP |
| Molecular Function: |
receptor signaling protein serine/threonine kinase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSEDEFFGGD NEAVWNGSRF SDSPEFQTLK EEVAAELFEI NGQISTLQQF TATLKSFIDR 60
61 GDVSAKVVER INKRSVAKIE EIGGLIKKVN TSVKKMDAIE EASLDKTQII AREKLVRDVS 120
121 YSFQEFQGIQ RQFTQVMKQV NERAKESLEA SEMANDAALL DEEQRQNSSK STRIPGSQIV 180
181 IERDPINNEE FAYQQNLIEQ RDQEISNIER GITELNEVFK DLGSVVQQQG VLVDNIEANI 240
241 YTTSDNTQLA SDELRKAMRY QKRTSRWRVY LLIVLLVMLL FIFLIMKL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
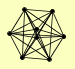
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
PEP12 SEC20 SED5 SSO1 SSO2 TLG1 TLG2 UFE1 |
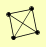
| View Details | Gavin AC, et al. (2006) |
|
PEP12 SEC17 VTI1 YKT6 |
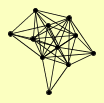
| View Details | Qiu et al. (2008) |
|
BET1 PEP12 SED5 SNC1 SNC2 SSO1 SSO2 TLG1 TLG2 VAM3 VTI1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | Sample: HX12 - trans-SNARE complex formation and fusion are inhibited (control). | Hao Xu, et al. (2010) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Uetz P, et al. (2000) | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
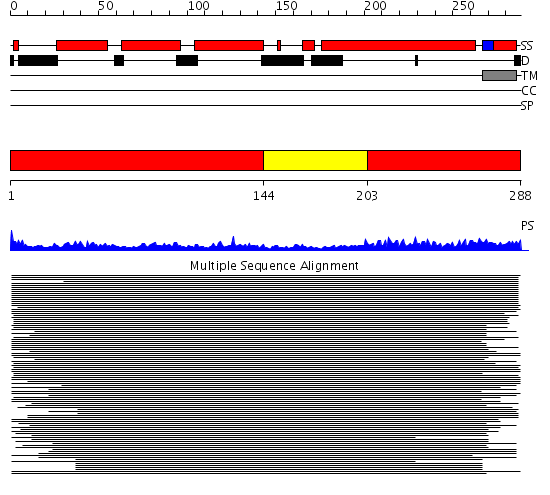
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..143] [203..288] |
86.0103 | Syntaxin 1A N-terminal domain | |
| 2 | View Details | [144..202] | 86.0103 | Syntaxin 1A N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|