













| General Information: |
|
| Name(s) found: |
TLG2 /
YOL018C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 397 amino acids |
Gene Ontology: |
|
| Cellular Component: |
early endosome
[TAS trans-Golgi network [TAS Golgi trans cisterna [TAS |
| Biological Process: |
vesicle fusion
[TAS |
| Molecular Function: |
receptor signaling protein serine/threonine kinase activity
[TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFRDRTNLFL SYRRTFPHNI TFSSGKAPLG DDQDIEMGTY PMMNMSHDIS ARLTDERKNK 60
61 HENHSDALPP IFIDIAQDVD DYLLEVRRLS EQLAKVYRKN SLPGFEDKSH DEALIEDLSF 120
121 KVIQMLQKCY AVMKRLKTIY NSQFVDGKQL SREELIILDN LQKIYAEKIQ TESNKFRVLQ 180
181 NNYLKFLNKD DLKPIRNKAS AENTLLLDDE EEEAAREKRE GLDIEDYSKR TLQRQQQLHD 240
241 TSAEAYLRER DEEITQLARG VLEVSTIFRE MQDLVVDQGT IVDRIDYNLE NTVVELKSAD 300
301 KELNKATHYQ KRTQKCKVIL LLTLCVIALF FFVMLKPHGG GSGGRNNGSN KYNNDDNKTV 360
361 NNSHDDGSNT HINDEESNLP SIVEVTESEN DALDDLL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
NAS6 TLG2 VPS45 |
| View Details | (MIPS) Mewes HW, et al. (2004) |
|
PEP12 SEC20 SED5 SSO1 SSO2 TLG1 TLG2 UFE1 |
| View Details | Krogan NJ, et al. (2006) |

|
ADE2 BSC2 CWH41 DBF4 DIG1 ECM29 HAL5 LAA1 MRC1 OPI9 PEX32 PRP5 PTH2 RGA2 RMA1 RPN3 SIR1 STE12 THR1 TLG2 VPS45 YDL199C YEL057C YPL035C |
| View Details | Gavin AC, et al. (2002) |
|
SEC17 SLY1 TLG1 TLG2 VPS45 VTI1 YKT6 |
| View Details | Qiu et al. (2008) |
|
DRS2 PEP12 SEC22 SED5 SNC1 SNC2 SSO1 SSO2 TLG1 TLG2 VTI1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Reinke A, et al. (2004) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | Sample: HX14 - mixture in detergent (control). | Hao Xu, et al. (2010) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data | ||
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
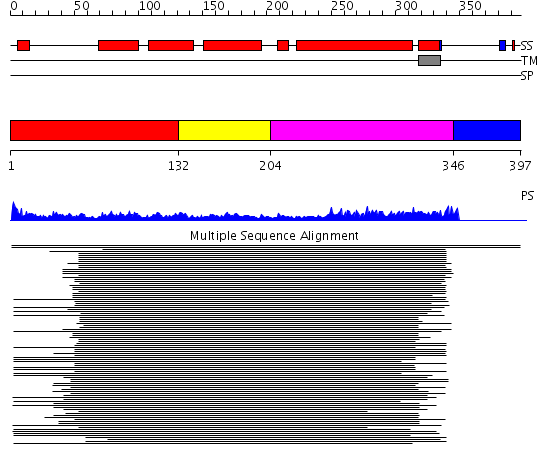
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..131] | 14.154902 | Syntaxin 1A N-terminal domain | |
| 2 | View Details | [132..203] | 14.154902 | Syntaxin 1A N-terminal domain | |
| 3 | View Details | [204..345] | 61.325697 | Syntaxin 1A | |
| 4 | View Details | [346..397] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|