













| General Information: |
|
| Name(s) found: |
PDR1 /
YGL013C
[SGD]
|
| Description(s) found:
Found 23 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1068 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IPI |
| Biological Process: |
response to drug
[IMP regulation of transcription from RNA polymerase II promoter [TAS |
| Molecular Function: |
DNA binding
[IPI transcription activator activity [TAS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRGLTPKNGV HIETGPDTES SADSSNFSTG FSGKIRKPRS KVSKACDNCR KRKIKCNGKF 60
61 PCASCEIYSC ECTFSTRQGG ARIKNLHKTS LEGTTVQVKE ETDSSSTSFS NPQRCTDGPC 120
121 AVEQPTKFFE NFKLGGRSSG DNSGSDGKND DDVNRNGFYE DDSESQATLT SLQTTLKNLK 180
181 EMAHLGTHVT SAIESIELQI SDLLKRWEPK VRTKELATTK FYPNKSIETQ LMKNKYCDVV 240
241 HLTRYAAWSN NKKDQDTSSQ PLIDEIFGLY SPFQFLSLQG IGKCFQNYRS KSKCEIFPRT 300
301 AKETIYIMLR FFDVCFHHIN QGCVSIANPL ENYLQKMNLL PSTPSSISSA GSPNTAHTKS 360
361 HVALVINHLP QPFVRNITGI SNSELLSEMN NDISMFGILL KMLDMHKNSY QNFLMEITSN 420
421 PSVAKNTQSI DVLQEFIHYC QAGEALIALC YSYYNSTLYN YVDFTCDITH LEQLLYFLDL 480
481 LFWLSEIYGF EKVLNVAVHF VSRVGLSRWE FYVGLDENFA ERRRNLWWKA FYFEKTLASK 540
541 LGYPSNIDDS KINCLLPKNF RDVGFLDNRD FIENVHLVRR SEAFDNMCIS DLKYYGELAV 600
601 LQIVSHFSSS VLFNEKFTSI RNTSKPSVVR EKLLFEVLEI FNETEMKYDA IKEQTGKLFD 660
661 IAFSKDSTEL KVSREDKIMA SKFVLFYEHH FCRMVNESDN IVARLCVHRR PSILIENLKI 720
721 YLHKIYKSWT DMNKILLDFD NDYSVYRSFA HYSISCIILV SQAFSVAEFI KVNDVVNMIR 780
781 VFKRFLDIKI FSENETNEHV FNSQSFKDYT RAFSFLTIVT RIMLLAYGES SSTNLDVISK 840
841 YIDENAPDLK GIIELVLDTN SCAYRFLLEP VQKSGFHLTV SQMLKNRKFQ EPLMSNEDNK 900
901 QMKHNSGKNL NPDLPSLKTG TSCLLNGIES PQLPFNGRSA PSPVRNNSLP EFAQLPSFRS 960
961 LSVSDMINPD YAQPTNGQNN TQVQSNKPIN AQQQIPTSVQ VPFMNTNEIN NNNNNNNNNK1020
1021 NNINNINNNN SNNFSATSFN LGTLDEFVNN GDLEDLYSIL WSDVYPDS |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |
|
CRP1 IDS2 PDR1 PEP4 PRC1 RTN1 SIA1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | cac1: TAP-tagged | Green EM, et al (2005) | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | identification data - first search in cascade | Green EM, et al (2005) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 2 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
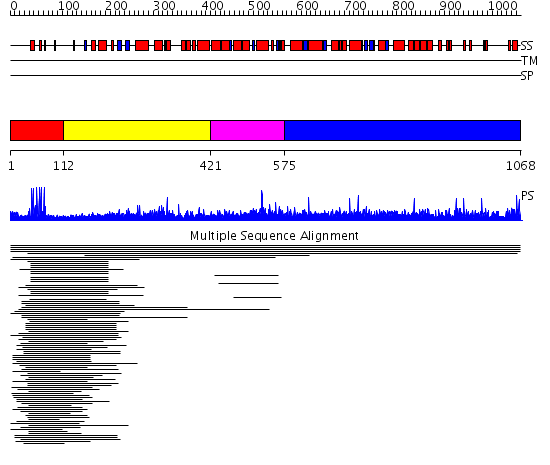
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] | 36.274545 | Gal4; CD2-Gal4 | |
| 2 | View Details | [112..420] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [421..574] | 3.012997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [575..1068] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [467..557] | 17.420216 | No description for PF04082.9 was found. No confident structure predictions are available. | |
| 6 | View Details | [558..798] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [799..1068] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)