













| General Information: |
|
| Name(s) found: |
CHD1 /
YER164W
[SGD]
|
| Description(s) found:
Found 25 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 1468 amino acids |
Gene Ontology: |
|
| Cellular Component: |
transcription elongation factor complex
[IPI SLIK (SAGA-like) complex [IDA mitochondrion [IDA SAGA complex [IDA |
| Biological Process: |
chromatin remodeling
[IDA RNA elongation from RNA polymerase II promoter [IPI |
| Molecular Function: |
ATPase activity
[IDA RNA polymerase II transcription elongation factor activity [IPI |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAAKDISTEV LQNPELYGLR RSHRAAAHQQ NYFNDSDDED DEDNIKQSRR KRMTTIEDDE 60
61 DEFEDEEGEE DSGEDEDEED FEEDDDYYGS PIKQNRSKPK SRTKSKSKSK PKSQSEKQST 120
121 VKIPTRFSNR QNKTVNYNID YSDDDLLESE DDYGSEEALS EENVHEASAN PQPEDFHGID 180
181 IVINHRLKTS LEEGKVLEKT VPDLNNCKEN YEFLIKWTDE SHLHNTWETY ESIGQVRGLK 240
241 RLDNYCKQFI IEDQQVRLDP YVTAEDIEIM DMERERRLDE FEEFHVPERI IDSQRASLED 300
301 GTSQLQYLVK WRRLNYDEAT WENATDIVKL APEQVKHFQN RENSKILPQY SSNYTSQRPR 360
361 FEKLSVQPPF IKGGELRDFQ LTGINWMAFL WSKGDNGILA DEMGLGKTVQ TVAFISWLIF 420
421 ARRQNGPHII VVPLSTMPAW LDTFEKWAPD LNCICYMGNQ KSRDTIREYE FYTNPRAKGK 480
481 KTMKFNVLLT TYEYILKDRA ELGSIKWQFM AVDEAHRLKN AESSLYESLN SFKVANRMLI 540
541 TGTPLQNNIK ELAALVNFLM PGRFTIDQEI DFENQDEEQE EYIHDLHRRI QPFILRRLKK 600
601 DVEKSLPSKT ERILRVELSD VQTEYYKNIL TKNYSALTAG AKGGHFSLLN IMNELKKASN 660
661 HPYLFDNAEE RVLQKFGDGK MTRENVLRGL IMSSGKMVLL DQLLTRLKKD GHRVLIFSQM 720
721 VRMLDILGDY LSIKGINFQR LDGTVPSAQR RISIDHFNSP DSNDFVFLLS TRAGGLGINL 780
781 MTADTVVIFD SDWNPQADLQ AMARAHRIGQ KNHVMVYRLV SKDTVEEEVL ERARKKMILE 840
841 YAIISLGVTD GNKYTKKNEP NAGELSAILK FGAGNMFTAT DNQKKLEDLN LDDVLNHAED 900
901 HVTTPDLGES HLGGEEFLKQ FEVTDYKADI DWDDIIPEEE LKKLQDEEQK RKDEEYVKEQ 960
961 LEMMNRRDNA LKKIKNSVNG DGTAANSDSD DDSTSRSSRR RARANDMDSI GESEVRALYK1020
1021 AILKFGNLKE ILDELIADGT LPVKSFEKYG ETYDEMMEAA KDCVHEEEKN RKEILEKLEK1080
1081 HATAYRAKLK SGEIKAENQP KDNPLTRLSL KKREKKAVLF NFKGVKSLNA ESLLSRVEDL1140
1141 KYLKNLINSN YKDDPLKFSL GNNTPKPVQN WSSNWTKEED EKLLIGVFKY GYGSWTQIRD1200
1201 DPFLGITDKI FLNEVHNPVA KKSASSSDTT PTPSKKGKGI TGSSKKVPGA IHLGRRVDYL1260
1261 LSFLRGGLNT KSPSADIGSK KLPTGPSKKR QRKPANHSKS MTPEITSSEP ANGPPSKRMK1320
1321 ALPKGPAALI NNTRLSPNSP TPPLKSKVSR DNGTRQSSNP SSGSAHEKEY DSMDEEDCRH1380
1381 TMSAIRTSLK RLRRGGKSLD RKEWAKILKT ELTTIGNHIE SQKGSSRKAS PEKYRKHLWS1440
1441 YSANFWPADV KSTKLMAMYD KITESQKK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
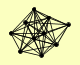
| View Details | Riffle et al. (2010) (Unpublished Data) |
|
CHD1 CKA1 CKA2 CKB1 CKB2 PAF1 POB3 RTF1 SPT16 UTP22 |
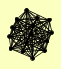
| View Details | Krogan NJ, et al. (2006) |
|
CAB3 CHD1 CKA1 CKA2 CKB1 CKB2 ELF1 HOT1 MSC3 NRG1 PGA2 REH1 RIM13 VHS3 YLR407W |
| View Details | Gavin AC, et al. (2006) |

|
CHD1 SSA4 |
| View Details | Gavin AC, et al. (2002) |

|
CAB3 CHD1 CKA1 CKA2 CKB1 CKB2 HHF1, HHF2 HOT1 ISW1 ISW2 NIP1 PRT1 RET1 RPG1 RVB2 SPT16 SSA4 TIF35 UTP22 VPS1 YTA7 |
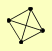
| View Details | Ho Y, et al. (2002) |
|
CHD1 HSH49 MLC2 RSE1 |
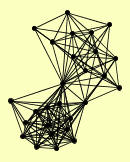
| View Details | Qiu et al. (2008) |
|
ADA2 ARP4 CDC73 CHD1 ELC1 ELP2 ELP6 GCN5 HFI1 IKI1 IKI3 LEO1 NGG1 PAF1 SGF73 SPN1 SPT16 SPT5 SPT6 SPT8 SUS1 TAF12 TAF14 TAF5 TAF6 TAF9 TFB5 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | No Comments | Toshima J, et al (2006) | |
| View Run | sample cbf3 from feb 2005 | Sandall S, et all (2006) | |
| View Run | #25 Asynchronous Prep3-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #26 Asynchronous Prep4-TiO2 Phosphopeptide enriched, Steps1-2 | Keck JM, et al. (2011) | |
| View Run | #29 Asynchronous Prep5-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #12 Mitotic Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) | |
| View Run | #02 Alpha Factor Prep1-TiO2 enriched, new search criteria | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Hazbun TR, et al. (2003) | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
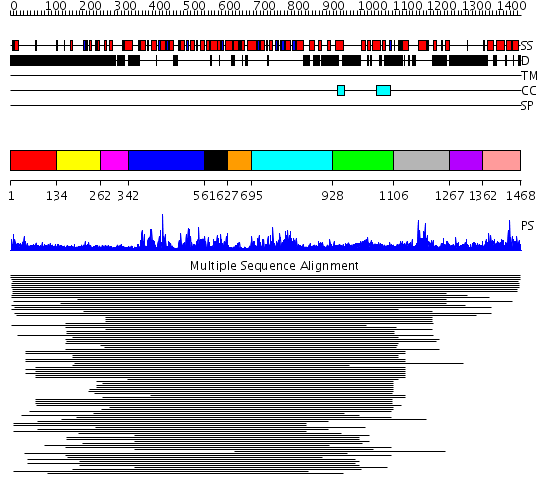
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [134..261] | 17.065502 | 'chromo' (CHRromatin Organisation MOdifier) domain No confident structure predictions are available. | |
| 3 | View Details | [262..341] | 13.61 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 4 | View Details | [342..560] | 13.61 | RecG, N-terminal domain; RecG "wedge" domain; RecG helicase domain | |
| 5 | View Details | [561..626] | 12.85 | Putative DEAD box RNA helicase | |
| 6 | View Details | [627..694] | 7.221849 | Putative DEAD box RNA helicase | |
| 7 | View Details | [695..927] | 7.30103 | Nucleotide excision repair enzyme UvrB | |
| 8 | View Details | [928..1105] | 1.175981 | View MSA. Confident ab initio structure predictions are available. | |
| 9 | View Details | [1106..1266] | 3.051976 | View MSA. No confident structure predictions are available. | |
| 10 | View Details | [1267..1361] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1362..1468] | 1.009974 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 9 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 10 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 11 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)