













| General Information: |
|
| Name(s) found: |
HEM13 /
YDR044W
[SGD]
|
| Description(s) found:
Found 36 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 328 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial inner membrane
[ISS |
| Biological Process: |
heme biosynthetic process
[TAS |
| Molecular Function: |
coproporphyrinogen oxidase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPAPQDPRNL PIRQQMEALI RRKQAEITQG LESIDTVKFH ADTWTRGNDG GGGTSMVIQD 60
61 GTTFEKGGVN VSVVYGQLSP AAVSAMKADH KNLRLPEDPK TGLPVTDGVK FFACGLSMVI 120
121 HPVNPHAPTT HLNYRYFETW NQDGTPQTWW FGGGADLTPS YLYEEDGQLF HQLHKDALDK 180
181 HDTALYPRFK KWCDEYFYIT HRKETRGIGG IFFDDYDERD PQEILKMVED CFDAFLPSYL 240
241 TIVKRRKDMP YTKEEQQWQA IRRGRYVEFN LIYDRGTQFG LRTPGSRVES ILMSLPEHAS 300
301 WLYNHHPAPG SREAKLLEVT TKPREWVK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
HEM13 MNL1 PDI1 |
| View Details | Gavin AC, et al. (2006) |
|
HEM13 PSK1 TAF7 VAS1 |
| View Details | Gavin AC, et al. (2002) |

|
ADA2 ADH1 CLU1 FUN19 GCN5 HEM13 HFI1 KAP114 KAP123 MED4 MES1 MNN4 MOT1 MYO1 MYO4 NGG1 NOP1 PFK2 PSK1 PTC1 PUF6 RPB2 RPN9 RPO21 RPT5 RVB2 SGF29 SGF73 SIN3 SIN4 SPT15 SPT20 SPT7 SPT8 TAF1 TAF10 TAF11 TAF12 TAF13 TAF14 TAF2 TAF3 TAF4 TAF5 TAF6 TAF7 TAF8 TAF9 TBF1 TEF4 UBP8 VAS1 VPS1 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
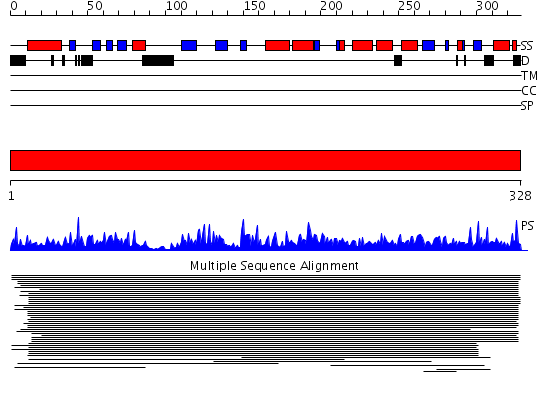
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..328] | 265.283997 | Coproporphyrinogen III oxidase No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)