













| General Information: |
|
| Name(s) found: |
MPK6_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 19 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 395 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
camalexin biosynthetic process
[IMP]
response to salt stress [IGI] signal transduction [IC] response to osmotic stress [IDA] induced systemic resistance, jasmonic acid mediated signaling pathway [IMP] priming of cellular response to stress [IMP] response to hydrogen peroxide [IDA] response to reactive oxygen species [IEP] defense response to bacterium [IEP] response to stress [TAS] response to ethylene stimulus [IDA] response to oxidative stress [IEP] response to cold [IDA] response to abscisic acid stimulus [IEP] ovule development [IGI] |
| Molecular Function: |
MAP kinase activity
[IDA][ISS]
kinase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDGGSGQPAA DTEMTEAPGG FPAAAPSPQM PGIENIPATL SHGGRFIQYN IFGNIFEVTA 60
61 KYKPPIMPIG KGAYGIVCSA MNSETNESVA IKKIANAFDN KIDAKRTLRE IKLLRHMDHE 120
121 NIVAIRDIIP PPLRNAFNDV YIAYELMDTD LHQIIRSNQA LSEEHCQYFL YQILRGLKYI 180
181 HSANVLHRDL KPSNLLLNAN CDLKICDFGL ARVTSESDFM TEYVVTRWYR APELLLNSSD 240
241 YTAAIDVWSV GCIFMELMDR KPLFPGRDHV HQLRLLMELI GTPSEEELEF LNENAKRYIR 300
301 QLPPYPRQSI TDKFPTVHPL AIDLIEKMLT FDPRRRITVL DALAHPYLNS LHDISDEPEC 360
361 TIPFNFDFEN HALSEEQMKE LIYREALAFN PEYQQ |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
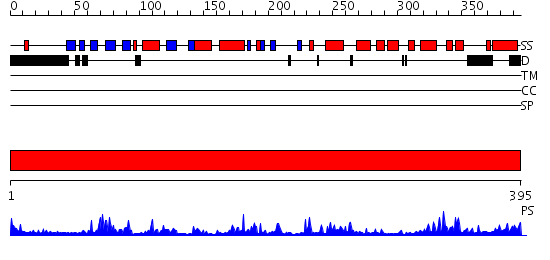
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..395] | 91.69897 | Crystal structure of P38 with triazolopyridine |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)