













| General Information: |
|
| Name(s) found: |
srk1 /
SPCC1322.08
[Sanger Pombe]
|
| Description(s) found:
Found 17 descriptions. SHOW ALL |
|
| Organism: | Schizosaccharomyces pombe |
| Length: | 580 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
cell cycle checkpoint
[IGI regulation of mitosis [IPI stress-activated protein kinase signaling pathway [IGI regulation of meiosis [IMP cellular response to stress [IEP protein amino acid phosphorylation [IDA |
| Molecular Function: |
ATP binding
[ISS protein binding [IPI calmodulin-dependent protein kinase activity [ISS protein serine/threonine kinase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRFKSIQQNI EDEGKVNVRE VNPDSYAERD HGYTAGIFSD AEENFGITQQ VADSTQNPTS 60
61 KPKSRHAHFH ETVHENPSEY SRSKCKQPTN EKEYDKAIEA LVAKAIVEEH SGQQFPVYKG 120
121 LEQYTLLQKM GDGAFSNVYK AIHNRTGEKV AIKVVQRAQP NTDPRDPRKR QGVESHNILK 180
181 EVQIMRRVKH PNIIQLLEFI QTPEYYYLVL ELADGGELFH QIVRLTYFSE DLSRHVITQV 240
241 AHAIRYLHED CGVVHRDIKP ENLLFDSIDF VPSRVRKYRA GDDPDKVDEG EFIPGVGAGT 300
301 IGRIRLADFG LSKVVWDSHT QTPCGTMGYT APEIVRDERY SKGVDMWALG CVLYTILCGF 360
361 PPFYDESISL LTKKVSRGEY SFLSPWWDDI SKSAKDLISH LLTVDPESRY DIHQFLAHPW 420
421 ISGSREPTFP ATDAPNTAQR ENPFTYDFLE PEDVAAAGSA ARTPGVNSLR EVFNISYAAH 480
481 RMEQEKIRKR GQRGNQGIMN FMGDMDDLME ENDDYDDGTK SVEHSMKRVN LSGENDPSLA 540
541 SRQPAQSQQQ SSQRSRNKFK GFQLNLSKAT LYNRRHRQKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
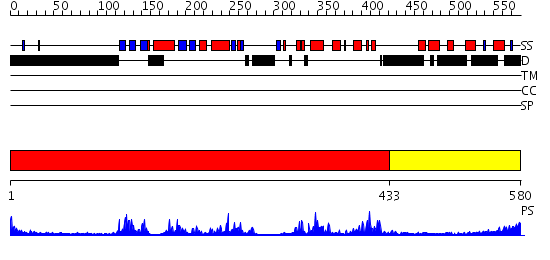
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..432] | 51.69897 | No description for 2fo0A was found. | |
| 2 | View Details | [433..580] | 66.0 | Crystal Structure of the Auto-Inhibited Kinase Domain of Calcium/Calmodulin Activated Kinase II |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)