













| General Information: |
|
| Name(s) found: |
FBpp0306059
[FlyBase]
slmb-PA / FBpp0083434 [FlyBase] |
| Description(s) found:
Found 32 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 510 amino acids |
Gene Ontology: |
|
| Cellular Component: |
ubiquitin ligase complex
[ISS]
SCF ubiquitin ligase complex [NAS][TAS] |
| Biological Process: |
negative regulation of nurse cell apoptosis
[IMP]
germarium-derived egg chamber formation [IMP] regulation of mitosis [IMP] regulation of proteolysis [TAS] release of cytoplasmic sequestered NF-kappaB [IMP][TAS] germarium-derived female germ-line cyst encapsulation [IMP] dorsal appendage formation [IMP] olfactory learning [IMP] border follicle cell migration [IMP] circadian rhythm [IMP][IPI] chorion-containing eggshell formation [IMP] negative regulation of smoothened signaling pathway [IMP] regulation of smoothened signaling pathway [TAS] learning or memory [IMP] ovarian follicle cell stalk formation [IMP] germarium-derived female germ-line cyst formation [IMP] locomotor rhythm [IMP] ovarian follicle cell development [IMP] proteolysis [IMP] negative regulation of Wnt receptor signaling pathway [NAS] regulation of Wnt receptor signaling pathway [IMP][TAS] negative regulation of transforming growth factor beta receptor signaling pathway [NAS] oocyte localization during germarium-derived egg chamber formation [IMP] |
| Molecular Function: |
ubiquitin-protein ligase activity
[ISS]
phosphoprotein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMKMETDKIM DETNSNAQAF TTTMLYDPVR KKDSSPTYQT ERELCFQYFT QWSESGQVDF 60
61 VEHLLSRMCH YQHGQINAYL KPMLQRDFIT LLPIKGLDHI AENILSYLDA ESLKSSELVC 120
121 KEWLRVISEG MLWKKLIERK VRTDSLWRGL AERRNWMQYL FKPRPGQTQR PHSFHRELFP 180
181 KIMNDIDSIE NNWRTGRHML RRINCRSENS KGVYCLQYDD GKIVSGLRDN TIKIWDRTDL 240
241 QCVKTLMGHT GSVLCLQYDD KVIISGSSDS TVRVWDVNTG EMVNTLIHHC EAVLHLRFNN 300
301 GMMVTCSKDR SIAVWDMTSP SEITLRRVLV GHRAAVNVVD FDEKYIVSAS GDRTIKVWST 360
361 SSCEFVRTLN GHKRGIACLQ YRDRLVVSGS SDNSIRLWDI ECGACLRVLE GHEELVRCIR 420
421 FDTKRIVSGA YDGKIKVWDL VAALDPRAAS NTLCLNTLVE HTGRVFRLQF DEFQIVSSSH 480
481 DDTILIWDFL NFTPNENKTG RTPSPALMEH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
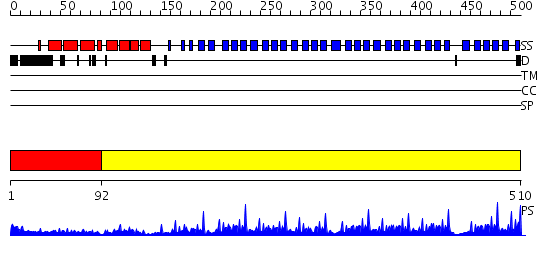
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..91] | 7.69897 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 2 | View Details | [92..510] | 97.69897 | F-box/WD-repeat protein 1 (beta-TRCP1) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)