













| General Information: |
|
| Name(s) found: |
FBpp0292491
[FlyBase]
FBpp0292487 [FlyBase] |
| Description(s) found:
Found 35 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 606 amino acids |
Gene Ontology: |
|
| Cellular Component: |
subapical complex
[TAS]
cytoplasm [NAS] cell cortex [TAS] apical cortex [IDA][NAS] apical plasma membrane [NAS][TAS] |
| Biological Process: |
cell-cell junction assembly
[NAS]
germarium-derived oocyte fate determination [IMP] establishment or maintenance of cell polarity [NAS] positive regulation of neuroblast proliferation [IMP] protein amino acid phosphorylation [IEA][NAS] memory [IMP] sensory organ development [IMP] oocyte anterior/posterior axis specification [IGI] establishment or maintenance of neuroblast polarity [NAS] oocyte axis specification [TAS] synapse assembly [IMP] intracellular signaling pathway [IEA] asymmetric protein localization involved in cell fate determination [IPI] zonula adherens assembly [NAS][TAS] establishment or maintenance of epithelial cell apical/basal polarity [IMP][NAS][TAS] apical protein localization [NAS] maintenance of cell polarity [IMP] morphogenesis of a polarized epithelium [TAS] asymmetric neuroblast division [IGI] establishment or maintenance of polarity of embryonic epithelium [NAS] |
| Molecular Function: |
ATP binding
[IEA]
zinc ion binding [IEA] protein binding [IPI][ISS] protein serine/threonine kinase activity [IEA][NAS] protein kinase C activity [NAS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQKMPSQILN DGSSVSLNSA SMNMANTPNS ITVKTAYNGQ IIITTINKNI SYEELCYEIR 60
61 NICRFPLDQP FTIKWVDEEN DPCTISTKME LDEAIRLYEM NFDSQLVIHV FPNVPQAPGL 120
121 SCDGEDRSIY RRGARRWRKL YRVNGHIFQA KRFNRRAFCA YCQDRIWGLG RQGFKCIQCK 180
181 LLVHKKCHKL VQKHCTDQPE PLVKERAEES SDPIPVPLPP LPYEAMSGGA EACETHDHAH 240
241 IVAPPPPEDP LEPGTQRQYS LNDFELIRVI GRGSYAKVLM VELRRTRRIY AMKVIKKALV 300
301 TDDEDIDWVQ TEKHVFETAS NHPFLVGLHS CFQTPSRLFF VIEFVRGGDL MYHMQRQRRL 360
361 PEEHARFYAA EISLALNFLH EKGIIYRDLK LDNVLLDHEG HIKLTDYGMC KEGIRPGDTT 420
421 STFCGTPNYI APEILRGEDY GFSVDWWALG VLLYEMLAGR SPFDLAGASE NPDQNTEDYL 480
481 FQVILEKTIR IPRSLSVRAA SVLKGFLNKN PADRLGCHRE SAFMDIVSHP FFKNMDWELL 540
541 ERKQVTPPFK PRLDSDRDLA NFPPEFTGEA VQLTPDDDHV IDNIDQSEFE GFEYVNPLLM 600
601 SLEDCV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
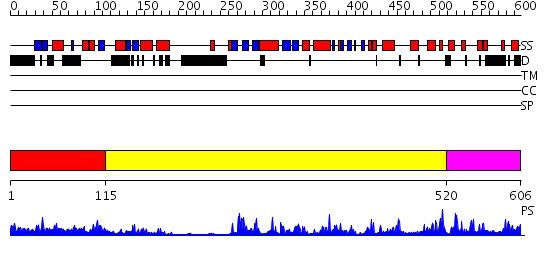
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..114] | 21.30103 | Solution Structure of the PB1 domain of PKCiota | |
| 2 | View Details | [115..519] | 50.522879 | CHICKEN SRC TYROSINE KINASE | |
| 3 | View Details | [520..606] | 94.09691 | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)