













| General Information: |
|
| Name(s) found: |
FBpp0307854
[FlyBase]
cos-PA / FBpp0088087 [FlyBase] |
| Description(s) found:
Found 28 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 1201 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IDA][TAS]
vesicle membrane [IDA] kinesin complex [ISS] Hedgehog signaling complex [IPI] microtubule associated complex [IPI] plasma membrane [IDA] |
| Biological Process: |
regulation of protein stability
[IDA]
ovarian follicle cell development [TAS] regulation of proteolysis [NAS] positive regulation of smoothened signaling pathway [TAS] imaginal disc-derived wing morphogenesis [IMP] smoothened signaling pathway [IMP][IPI][TAS] negative regulation of transcription factor import into nucleus [IMP] negative regulation of transcription factor activity [IMP] cytoplasmic sequestering of transcription factor [IMP] microtubule-based movement [IEA][ISS] positive regulation of hh target transcription factor activity [IMP] negative regulation of smoothened signaling pathway [TAS] |
| Molecular Function: |
protein kinase binding
[IPI]
ATP binding [IEA] microtubule motor activity [IEA][ISS] protein binding [IPI] smoothened binding [IPI] transcription factor binding [IPI] microtubule binding [IDA] motor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEIPIQVAVR IFPHRELKDL LRSFGPTEPK KDAQAVDEGA DSKDSEAQVP AAEKDNPSIS 60
61 ETDPNGNAEQ DSAADSKTIP DANGNDSGQK DYPDSAYCVQ AIPISASALG LPSALPGGDP 120
121 MDSIAAGLIQ VGPHTVPVTH ALPSSSSQEQ VYHQTVFPLI TLFLEGFDAS VVTYGQRGQG 180
181 KSYTLYGNVQ DPTLTDSTEG VVQLCVRDIF SHISLHPERT YAINVGFVEI CGGDVCDLLG 240
241 MGNIHCTNVD AVFHWLQVGL SARQSLPAHT LFTLTLEQQW VSKEGLLQHR LSTASFSDLC 300
301 GTERCGDQPP GRPLDAGLCM LEQVISTLTD PGLMYGVNGN IPYGQTTLTT LLKDSFGGRA 360
361 QTLVILCVSP LEEHLPETLG NLQFAFKVQC VRNFVIMNTY SDDNTMIVQP AEPVPESNSS 420
421 AGPLSQAGPG DNFGLQFAAS QWSKLVTNAE GLFSKLIDSK LITEVEKEQI EEWLFLKQEC 480
481 EECLSSTEAM RQQKQLVPIL EAEEPEDVNS EAANSESPNS DNENDTDNES HRPDLDDKIE 540
541 SLMEEFRDKT DALILEKHAE YLSKHPKAVM QSQDREIEAQ PPEENGDDRK VSIGSRRRSV 600
601 QPGASLSTAE LAMLNRVASQ QPPPPIDPES VVDPLESSSG EGIRQAALAA AAATAPIEQL 660
661 QKKLRKLVAE IEGKQRQLRE IEETIQVKQN IIAELVKNSD TRSHAKQRFH KKRAKLEAEC 720
721 DKAKKQLGKA LVQGRDQSEI ERWTTIIGHL ERRLEDLSSM KHIAGESGQK VKKLQQSVGE 780
781 SRKQADDLQK KLRKECKLRC QMEAELAKLR ESRETGKELV KAQGSPEQQG RQLKAVQARI 840
841 THLNHILREK SDNLEEQPGP EQQETLRHEI RNLRGTRDLL LEERCHLDRK LKRDKVLTQK 900
901 EERKLLECDE AIEAIDAAIE FKNEMITGHR SIDTSDRIQR EKGEQMLMAR LNRLSTEEMR 960
961 TLLYKYFTKV IDLRDSSRKL ELQLVQLERE RDAWEWKERV LSNAVRQARL EGERNAVLLQ1020
1021 RQHEMKLTLM LRHMAEETSA SSASYGERAL APACVAPPVQ ASSDFDYDHF YKGGGNPSKA1080
1081 LIKAPKPMPT GSALDKYKDK EQRSGRNIFA KFHVLTRYAS AAAAGSSGST AEESTALIES1140
1141 TTTATATTTS TTTTGAVGKV KDKALVSFRP EQLKRLMPAP TATKVTRQKN KIIIQDASRR1200
1201 N |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
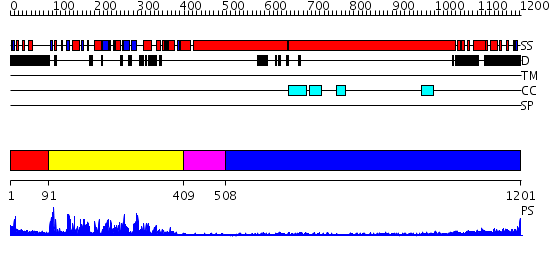
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..90] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [91..408] | 87.30103 | Kinesin | |
| 3 | View Details | [409..507] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [508..1201] | 14.522879 | Heavy meromyosin subfragment |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)