













| General Information: |
|
| Name(s) found: |
PMT4 /
YJR143C
[SGD]
|
| Description(s) found:
Found 29 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 762 amino acids |
Gene Ontology: |
|
| Cellular Component: |
endoplasmic reticulum
[TAS |
| Biological Process: |
protein amino acid O-linked glycosylation
[IMP |
| Molecular Function: |
dolichyl-phosphate-mannose-protein mannosyltransferase activity
[IMP |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVPKKRNHG KLPPSTKDVD DPSLKYTKAA PKCEQVAEHW LLQPLPEPES RYSFWVTIVT 60
61 LLAFAARFYK IWYPKEVVFD EVHFGKFASY YLERSYFFDV HPPFAKMMIA FIGWLCGYDG 120
121 SFKFDEIGYS YETHPAPYIA YRSFNAILGT LTVPIMFNTL KELNFRAITC AFASLLVAID 180
181 TAHVTETRLI LLDAILIISI AATMYCYVRF YKCQLRQPFT WSWYIWLHAT GLSLSFVIST 240
241 KYVGVMTYSA IGFAAVVNLW QLLDIKAGLS LRQFMRHFSK RLNGLVLIPF VIYLFWFWVH 300
301 FTVLNTSGPG DAFMSAEFQE TLKDSPLSVD SKTVNYFDII TIKHQDTDAF LHSHLARYPQ 360
361 RYEDGRISSA GQQVTGYTHP DFNNQWEVLP PHGSDVGKGQ AVLLNQHIRL RHVATDTYLL 420
421 AHDVASPFYP TNEEITTVTL EEGDGELYPE TLFAFQPLKK SDEGHVLKSK TVSFRLFHVD 480
481 TSVALWTHND ELLPDWGFQQ QEINGNKKVI DPSNNWVVDE IVNLDEVRKV YIPKVVKPLP 540
541 FLKKWIETQK SMFEHNNKLS SEHPFASEPY SWPGSLSGVS FWTNGDEKKQ IYFIGNIIGW 600
601 WFQVISLAVF VGIIVADLIT RHRGYYALNK MTREKLYGPL MFFFVSWCCH YFPFFLMARQ 660
661 KFLHHYLPAH LIACLFSGAL WEVIFSDCKS LDLEKDEDIS GASYERNPKV YVKPYTVFLV 720
721 CVSCAVAWFF VYFSPLVYGD VSLSPSEVVS REWFDIELNF SK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ACK1 CIN8 DCG1 GNA1 MCM6 MLP1 PMD1 PMT4 PRE1 PRE10 PRE2 PRE3 PRE4 PRE5 PRE6 PRE7 PRE8 PRE9 PUP1 PUP2 PUP3 RFT1 SCL1 SGA1 SNQ2 TOS8 UMP1 YCR076C YDR179W-A YJL132W YKR070W YLR211C YLR290C |
| View Details | Qiu et al. (2008) |
|
ALG2 PMT1 PMT2 PMT3 PMT4 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | No Comments | Schneider, DA, et al. (2006) | |
| View Run | #32 Asynchronous Prep-No Phosphopeptide enrichment | Keck JM, et al. (2011) |
SHOWING SINGLE HITS. [ Hide Single Hits ]
| BAIT | PREY | HITS | PUBLICATION | |
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Unpublished Fields Lab Data | ||
| View Screen | 1 | Hazbun TR, et al. (2003) |
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
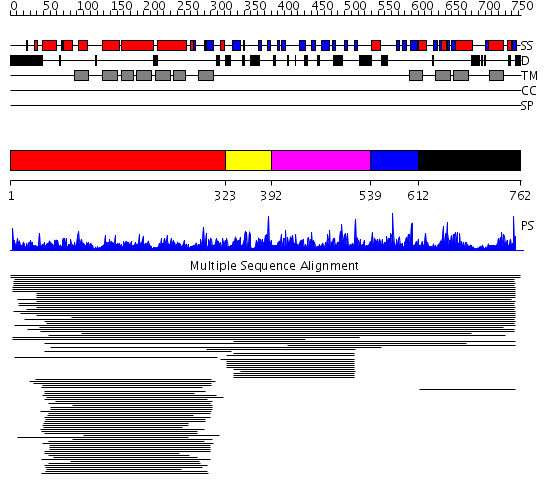
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..322] | 98.251812 | Dolichyl-phosphate-mannose-protein mannosyltransferase No confident structure predictions are available. | |
| 2 | View Details | [323..391] | 11.79588 | MIR domain No confident structure predictions are available. | |
| 3 | View Details | [392..538] | 10.63 | IP3 receptor type 1 binding core, domain 2; IP3 receptor type 1 binding core, domain 1 | |
| 4 | View Details | [539..611] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [612..762] | 1.028973 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|